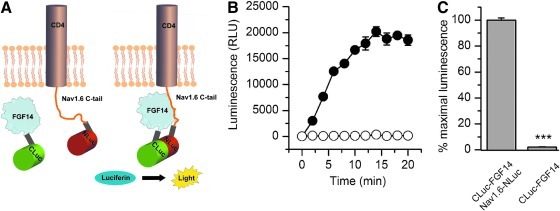
Fig. 1.
Bioluminescence detection of the FGF14:Nav1.6 C-tail complex in live cells using the split-luciferase complementation assay. (A) Schematic of the split-luciferase complementation assay (LCA). Constructs expressing the CLuc (398–550) and NLuc (2-415) fragments of firefly Photinus pyralis luciferase were fused, respectively, to full-length FGF14 and a chimera of the transmembrane protein CD4 and the C-terminal tail of Nav1.6. A flexible linker (gray) spaces the FGF14 and CD4-Nav1.6 cDNA from the two halves of luciferase. Spontaneous association of FGF14 and Nav1.6 C-tail brings in close proximity the two halves of luciferase leading to recombination of the luciferase enzymatic activity and luminescence production in the presence of the substrate D-luciferin (right). (B) HEK293 cells were transiently transfected with either CLuc-FGF14 or CD4-Nav1.6-NLuc (•) or CLuc-FGF14 alone (○). The assembly of the FGF14:Nav1.6 channel C-tail complex is detected as luminescence (relative luminescence units, RLU) upon the addition of the D-luciferin (0.75 mg/mL) substrate at time zero; data are mean±SEM from quadruplicate wells from one single experiment. (C) Bar graph represents % maximal bioluminescence measured upon functional complementation of the indicated constructs. The CLuc-FGF14+CD4-Nav1.6-NLuc pair represents the positive control, whereas CLuc-FGF14 alone serves as reference luminescence background. The data are mean±SEM representing eighth replicates from four independent experiments (CLuc-FGF14+CD4-Nav1.6-NLuc, n=4) or four replicates from three independent experiments (CLuc-FGF14 alone, n=3). The background level was 2.1±0.25% compared to the positive control. The signal-to-background level was 81±4.2. The mean values are compared using Student's t-test; ***p<0.001.