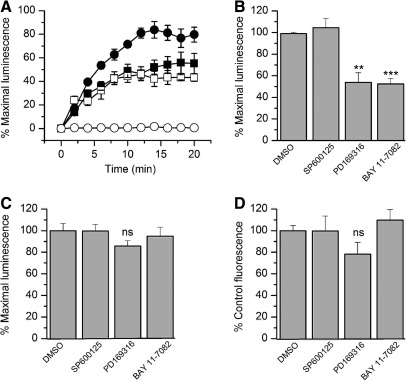
Fig. 4.
The FGF14:Nav1.6 functional complementation is regulated by specific kinase inhibitors. (A) HEK293 cells were transiently transfected with CLuc-FGF14 and CD4-Nav1.6-NLuc and treated with the p38 kinase inhibitor PD169316 (10 μM; ▪), the IKK inhibitor, BAY 11-7082 (10 μM; □) or dimethylsulfoxide (DMSO; 0.5% control; •). Cells transfected with CLuc-FGF14 alone (○) are also shown. Assembly of the LCA pair is detected as luminescence upon the addition of the D-luciferin substrate at time zero and normalized to % maximal luminescence signal in DMSO (0.5%, control); data are mean±SEM, representing quadruplicates from one representative experiment. (B) Bar graph represents mean±SEM expressed as % maximal luminescence of control (0.5% DMSO). The graphs illustrates the effect of the c-JNK inhibitor SP600125 (50 μM), the p38 kinase inhibitor PD169316 (10 μM), and the IKK inhibitor BAY 11-7082 (10 μM) on the FGF14:Nav1.6 channel C-tail assembly. The control and the SP600125 and PD169316 experimental groups each consisted of four replicates from four independent experiments (n=4); the BAY 11-7082 experimental group consisted of four replicates from three independent experiments (n=3). Statistical significance between the four groups was assessed using one-way ANOVA, post hoc Dunnett's method, **p<0.01, ***p<0.001. (C) HEK293 cells were transiently transfected with full-length firefly luciferase holoenzyme and treated for 1 h prior to the assay with the indicated compounds. Bar graph expressed as % maximal luminescence illustrates the effect of indicated compounds on the intrinsic enzymatic activity of luciferase; data are mean±SEM representing four replicates from three independent experiments (n=3). Statistical significance between the four groups was assessed using Kruskal–Wallis one-way ANOVA on ranks, post hoc Dunn's method; ns=nonsignificant. (D) HEK293 cells were transiently transfected with CLuc-FGF14 and CD4-Nav1.6-NLuc and treated with the indicated compounds. The effect of compounds on cell viability was determined by using CyQUANT® Cell Proliferation Assay Kit. Bar graph expressed as % control fluorescence illustrates the effect of the indicated compounds on cell viability. Data are mean±SEM, n=3. Statistical significance between the four groups was assessed using one-way ANOVA; ns=nonsignificant. None of the treatments in (D) were significantly different from each others, and no further post hoc analysis was required.