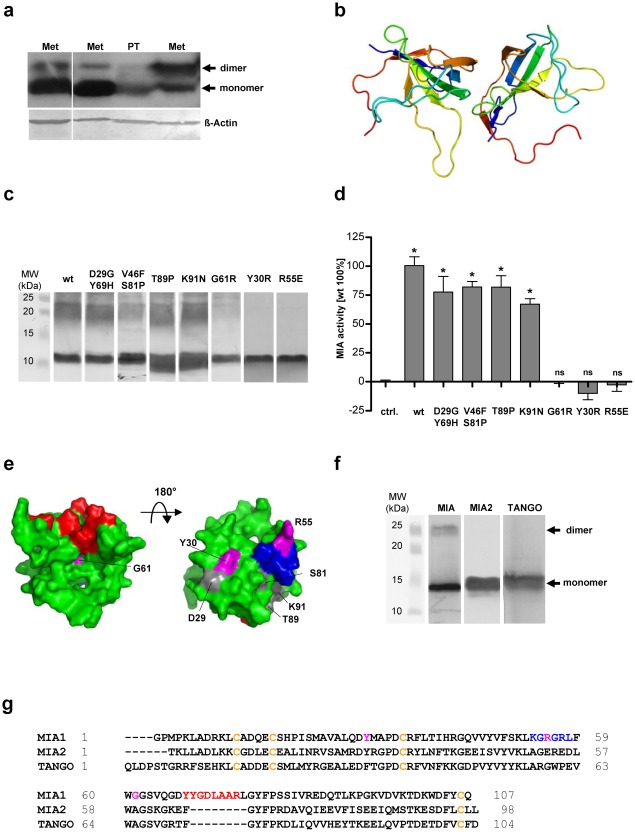
Figure 1. MIA is functionally active as a dimer.
(a) Western blot analysis of MIA in lysates from melanoma tissue (PT: primary tumor; Met: metastasis) under denaturating conditions. (b) The structure of the MIA dimer according to shape complementarity analyses. The MIA dimer is characterized by a head-to-tail orientation, with the dimerization domains consisting of the n-Src loop and the cleft next to the distal loop. (c) Western blot analysis of MIA mutants assessing their ability to form dimers. The first lane shows wt MIA, followed by the D29G/Y69H, V46F/S81P, T89P, K91N, G61R, Y30R and R55E mutants. All proteins except for G61R, Y30R and R55E have a clear dimer band. All proteins were electrophoretically resolved from an RTS expression system by SDS-PAGE. (d) The correlation between dimerization and functional activity revealed that all MIA mutants capable of dimerization are functionally active in Boyden chamber invasion assays. The G61R, Y30R and R55E mutants which do not form protein dimers, displayed no MIA-induced effect. (e) NMR structure of MIA showing the dimerization domains and the mutation sites. The dimerization domains in the n-Src loop and next to the distal loop are depicted in blue and red, respectively. The mutation sites that did not influence dimerization and functional activity are shown in gray. Residues Y30, R55 and G61 are shown in magenta. This figure was generated using PyMol [27]. (f) Western blot analysis of MIA and the MIA-homologous MIA2 and TANGO. Only MIA revealed dimerization. (g) Sequence alignment of MIA, the N-terminal part of MIA2 and TANGO. Conserved cysteines are shown in yellow. Residues important for MIA dimerization are shown in blue and red. The Y30, R55 and G61 mutation sites are shown in magenta (ns: not significant, *: p<0.05).