Abstract
Even skipped (Eve) and Engrailed (En) are homeodomain-containing transcriptional repressors with similar DNA binding specificities that are sequentially expressed in Drosophila embryos. The sloppy-paired (slp) locus is a target of repression by both Eve and En. At blastoderm, Eve is expressed in 7 stripes that restrict the posterior border of slp stripes, allowing engrailed (en) gene expression to be initiated in odd-numbered parasegments. En, in turn, prevents expansion of slp stripes after Eve is turned off. Prior studies showed that the two tandem slp transcription units are regulated by cis-regulatory modules (CRMs) with activities that overlap in space and time. An array of CRMs that generate 7 stripes at blastoderm, and later 14 stripes, surround slp1 (Fujioka and Jaynes, 2012). Surprisingly given their similarity in DNA binding specificity and function, responsiveness to ectopic Eve and En indicates that most of their direct target sites are either in distinct CRMs, or in different parts of coregulated CRMs. We localized cooperative binding sites for En, with the homeodomain-containing Hox cofactors Extradenticle (Exd) and Homothorax (Hth), within two CRMs that drive similar expression patterns. Functional analysis revealed two distinct, redundant sites within one CRM. The other CRM contains a single cooperative site that is both necessary and sufficient for repression in the en domain. Correlating in vivo and in vitro analysis suggests that cooperativity with Exd and Hth is a key ingredient in the mechanism of En-dependent repression, and that apparent affinity in vitro is an unreliable predictor of in vivo function.
Keywords: homeodomain protein binding, cooperative homeoprotein sites, Engrailed, Extradenticle, Homothorax, sloppy paired
Introduction
DNA-binding transcription factors function coordinately to regulate genes in eukaryotes. In many cases, this coordination begins with cooperative DNA binding to multiple sites within cis-regulatory modules or enhancers (Carey, 1998; Datta and Small, 2011). The actions of combinations of these proteins are further coordinated through the regulation of a gene by multiple enhancers, which are often active in patterns that overlap in space and time within an organism. This leads to apparent redundancy, which has been shown to lend robustness to the ability of some genes to provide full function in a variety of circumstances, such as a stressful environment (Frankel et al., 2010; Perry et al., 2010). Redundancy is also apparent in the action of related genes, which can sometimes partially substitute for each other. Thus, apparent redundancy can be seen at all levels of genomic organization, and its role in the evolution of gene regulation is just beginning to be elucidated.
The sloppy-paired (slp) locus contains two partially redundant, tandem transcription units, slp1 and slp2, that both encode transcription factors with a forkhead domain (Grossniklaus et al., 1992). During segmentation of the germ band, they act downstream of primary pair-rule genes such as even skipped (eve) (Fujioka et al., 1995), and so are secondary pair-rule genes (Cadigan et al., 1994). They help to establish and maintain parasegment (PS) boundaries, a conserved function (Choe and Brown, 2007). This occurs through slp expression in 7 and then 14 stripes, which are located on the anterior side of each PS boundary within the germ band. The engrailed (en) gene is expressed in 14 stripes just posterior to each PS boundary. The PS boundary is stabilized in part through mutual repression between en and slp (Cadigan et al., 1994; Jaynes and Fujioka, 2004; Kobayashi et al., 2003).
Both eve and en encode conserved homeodomain-containing transcriptional repressors (Akam, 1987; Fujioka et al., 2003; Fujioka et al., 2002; Jaynes and O’Farrell, 1991; Macdonald et al., 1986; Tolkunova et al., 1998; Zhang et al., 2002). In Drosophila, eve is expressed at blastoderm in 7 broad (“early”) stripes that are centered on the future odd-numbered PSs. Later, during gastrulation, its expression becomes restricted to the anterior-most cell rows of odd-numbered PSs (the “late” eve stripes), while the early 7-stripes of slp form just anterior to these stripes (Grossniklaus et al., 1992), so that the borders between them prefigure the anterior boundaries of the odd-numbered PSs (Frasch et al., 1987; Macdonald et al., 1986). En stripes are activated within eve stripes, through a well-studied mechanism that involves repression of slp. Briefly, the early bell-shaped concentration gradient within each early Eve stripe represses target genes in a concentration-dependent manner. The gene paired, an activator of En, is repressed only at high concentrations of Eve, while slp, a repressor of En, is repressed at both high and low Eve concentrations. This differential sensitivity creates a cell row that contains the activator (paired) but not the repressor (slp), turning en on in the anterior-most cell row of each odd-numbered PS (Fujioka et al., 1995). Repression by Runt prevents this from also happening at the posterior edge of early eve stripes (Ingham and Gergen, 1988; Jaynes and Fujioka, 2004; Manoukian and Krause, 1993). In the absence of eve, expanded slp expression prevents activation of en in these cells, even though paired is present (Frasch et al., 1988; Jaynes and Fujioka, 2004; Riechmann et al., 1997). Once en and slp expression are established, mutual repression between them helps to maintain the PS boundary (Cadigan et al., 1994; Kobayashi et al., 2003), which then serves as a signaling center for further cell specification within each PS. Therefore, slp is a key target gene for repression by both Eve and En.
Most homeodomain transcription factors show little sequence discrimination for DNA binding sites when assayed alone, and are thought to achieve sufficient specificity to carry out their distinct functions by binding cooperatively to target genes with other DNA binding proteins. While such cofactors for Eve have not been found, some cofactors for En have been identified. En has been shown to cooperate with Extradenticle (Exd) and Homothorax (Hth) to repress slp (Kobayashi et al., 2003) and distalless (Gebelein et al., 2004). Exd was identified as a cofactor of Hox proteins (Peifer and Wieschaus, 1990) (reviewed in (Mann et al., 2009)), while Hth dimerizes with Exd and induces its nuclear localization (Rieckhof et al., 1997).
Motivated by a desire to understand how Eve, En and cofactors regulate slp, we previously conducted a systematic analysis of the entire slp1 – slp 2 locus (Fujioka and Jaynes, 2012), and identified a 15 kilobase (kb) stripe-forming regulatory region surrounding the slp1 transcription unit. Within this region, a series of minimal stripe CRMs were localized by transgenic dissection.
Here, we identify functional binding regions for Eve and En among these slp stripe CRMs using ectopic expression assays in vivo. We find two distinct 14-stripe CRMs that are responsive to En, and which contain sites that mediate cooperative binding of En with Exd and Hth. Within each of these is a well-conserved En/Exd/Hth cooperative site accompanied by one or more less conserved sites. We find that one of the well-conserved sites is essential for repression by En within the context of its CRM, while the other is redundant with less conserved sites. Both of the well-conserved sites and one of the less conserved sites can substitute in vivo for a repression element that contains the essential site. Thus, even within the framework of cooperative binding with a specific set of cofactors, there is more than one way that binding sites are configured to achieve the same outcome. The site that does not confer repression shows a similar affinity, but less cooperativity, than the less conserved site that confers repression. This suggests that the degree of cooperativity, in addition to apparent affinity in vitro, may help predict in vivo occupancy.
Materials and Methods
Plasmids construction and production of transgenic flies
The plasmids and transgenic lines used for heat shock analysis were described previously (Fujioka and Jaynes, 2012). Briefly, CRM-driven transgenes used slp1 promoter-lacZ, in which the region from −261 to +121 bp relative to the slp1 transcription start site (TSS) was fused to the lacZ coding region followed by the eve 3′ UTR from +1306 to +1521 bp (KpnI) relative to the eve TSS. CRMs are inserted upstream. Analysis of u4734 and derivatives was done using standard P-element transgenesis. Several independent insertions were analyzed for each construct, and the expression patterns shown were consistently seen. To compare modifications of CRM i1523, and repression activity of En/Exd/Hth binding sites with CRM u4739, ΦC31 recombinase-mediated cassette exchange (ΦC31-RMCE) was used (Bateman et al., 2006). The various CRMs were cloned into attBΔ2 (Fujioka et al., 2008). Details of cloning procedures are available on request. ΦC31-RMCE was performed as previously described (Bateman et al., 2006; Groth et al., 2004), except that chromosomally integrated ΦC31 recombinase (Bischof et al., 2007) was used, instead of co-injection of ΦC31 mRNA. Successful RMCE events were first identified by loss of mini-white-dependent eye color. The presence and direction of the exchanged region were determined by PCR. The attP-docking site at cytological location 95E5 (Fujioka et al., 2008) was used, because it gave expression patterns for CRMs i1523, u4734, and u4739 that were indistinguishable from P-element insertions of the same constructs previously analyzed. In addition to 95E5, an attP-docking site at cytological location 23C1 was used for analysis of CRM i1523 modifications. Both docking sites gave the same results.
Embryo analysis
Embryos were subjected to in situ hybridization using anti-sense RNA probes against lacZ or en mRNA. The DIG-labeled probe was visualized by alkaline phosphatase-conjugated anti-DIG antibody (Roche Applied Science). For fluorescent in situ hybridization (FISH), DIG-labeled or Biotin-labeled probes (Jackson ImmunoResearch) were visualized with anti-DIG or anti-Biotin followed by DyLight549-conjugated anti-mouse IgG (Jackson ImmunoResearch) and DyLight488-conjugated anti-sheep IgG as described previously (Kosman et al., 2004). For double antibody staining, anti-βgalactosidase (ICN) and anti-En3 (4D9 monoclonal) obtained from the Developmental Studies Hybridoma Bank were visualized with DyLight549-conjugated anti-mouse IgG and DyLight488-conjugated anti-rabbit IgG (Jackson ImmunoResearch).
For quantifying repression activities of binding sites in the context of lacZ transgenes, embryos were first subjected to in situ hybridization with DIG-labeled lacZ mRNA probe, then to anti-En antibody staining (Santa Cruz Biotechnology) followed by biotin-conjugated anti-Rabbit IgG (Jackson ImmunoResearch), peroxidase-conjugated streptavidin (Jackson ImmunoResearch), and the DAB reaction. En-expressing cells were categorized into three classes: (−) no lacZ expression, (+) weak lacZ expression (weaker than lacZ-expressing cells outside the En domain), and (++) strong lacZ expression (similar to lacZ-expressing cells outside the En domain). Several stage 10–11 embryos were analyzed for each transgenic line.
Ectopic protein expression
Embryos carrying one copy of a transgene that expresses either En (hs-En, a.k.a. pRK232; (Heemskerk et al., 1991)) or Eve (hs-FLAG-Eve) in response to heat shock, along with a CRM-lacZ reporter transgene, were aged for 2.5–5.5 hr at room temperature (RT) after egg laying, dechorionated, heat-shocked for 10 min in a 37°C water bath, incubated for 30 min at RT, then fixed and stained for lacZ reporter mRNA. The hs-FLAG-eve transgene contains the entire Eve coding sequence with an added Flag tag at the 5′ end (see Fig. S1 for 5′ junction sequence), inserted into pCaSpeR-hs (Struhl, 1985). In each experiment, more than 500 embryos were analyzed. Control yw flies (the host line for transgenic production) were also crossed with each lacZ line, and processed in parallel. As an internal staining control, fixed embryos expressing a non-relevant pattern of lacZ were added to each set of heat-shocked, fixed embryos. Samples with closely comparable control staining intensity were compared to determine their relative responsiveness to En or Eve.
En/Exd/Hth binding site search criteria
Priority was given in choosing sequences for in vitro analysis within En-responsive CRMs based on the following criteria: a combination of an Exd consensus core (ATCA) and an En consensus core (ATTA) within 6 nt of each other, irrespective of relative orientation; or, in some cases, an Exd core with something like an En core (ATTG) in close proximity. Secondary priority was given to sequences containing a Hth core (TGAC, or [T,G]TGTC[A,C]) near an Exd core.
Electromobility shift assays
For use in electromobility shift assays (EMSAs) with radioactive probes, Exd and Hth proteins were purified from BL21 bacteria as His-tagged heterodimers using Ni-chromatography as previously described (Gebelein et al., 2002). A full-length His-tagged Engrailed protein was similarly purified under native conditions (50mM Tris, pH 7.5; 100mM NaCl; 0.1% NP-40; 10% glycerol). Protein concentrations were measured by the Bradford assay and confirmed by SDS-polyacrylamide gel electrophoresis followed by Coomassie blue analysis. EMSAs were performed as previously described (Uhl et al., 2010) using the following protein concentrations: 40 ng Exd/Hth per lane as indicated in Figs. 3D, 3E, and 6D, and 160 ng in Fig. 6E; for En, 25 (low), 75 (medium), and 225 (high) ng in Fig. 3D; 25 (low) and 225 (high) ng in Figs. 3E and 6D, and 50 ng in Fig. 6E. In all these cases, EMSAs within the same figure panel were performed at the same time and with the same protein concentrations and thus are directly comparable. For use in EMSAs with non-radioactive probes, His-tagged proteins were expressed in bacteria using pET15-EnΔS (codons 1–166 of En removed), pET15-Exd, and pET14b-Hth, purified according to the manufacturer’s protocol (His.Bind Purification Kit, EMD), and visualized by Coomassie blue staining and by ECL Western blot with anti-His monoclonal antibody (Covance). Exd and Hth were used at a 1:1 molar ratio based on the anti-His Western blot signal, since both proteins contain an N-terminal His-tag. Amounts of En and Exd/Hth were optimized based on cooperative binding to either A1 or B1 (see figures). Detailed protocols are available on request.
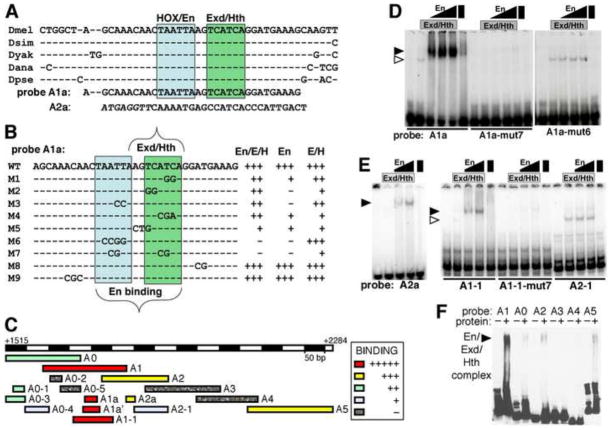
Fig. 3. Cooperative binding by Engrailed. Extradenticle and Homothorax to En-responsive CRM i1523.
A: conserved consensus sites for En (blue box), Exd and Hth (green box) located near each other within i1523.
B: summary of in vitro binding to probe A1a and mutated versions. Indicated to the right of each probe sequence is the strength of complex formation by En+Exd+Hth (“En/E/H”), En alone, or Exd+Hth (“E/H”) relative to that for A1a (arbitrarily set at “+++”) as determined by EMSA (see Materials and Methods). Examples (WT, M6 and M7) are shown in D.
C: summary of in vitro binding to probes from region A by En+Exd+Hth. Relative strength of binding in EMSAs is indicated by colors, listed in the inset. Examples are shown in D–F.
D: binding to probe A1a and mutant versions 6 and 7 (as indicated at the bottom, M6 and M7 in B, respectively) by Exd+Hth, En, and a combination of all 3, at 3 different concentrations of En (as indicated at the top, see Materials and Methods). The position of the main cooperative complex seen with all 3 proteins (which comigrates with the En complex) is indicated by a filled arrowhead, and the position of the Exd+Hth complex by an open arrowhead. Note that binding to A1a by all 3 proteins is cooperative, and that there is virtually no cooperative complex with either mutant version.
E: binding to other probes in region A (diagrammed in C, as indicated at the bottom) by Exd+Hth, En and a combination of all 3, at 2 different concentrations of En (as indicated at the top). The position of the main cooperative complex seen with all 3 proteins is indicated by a filled arrowhead, and the position of the Exd+Hth complex by an open arrowhead. Note that binding to A2a by all 3 proteins is cooperative, but weaker than binding to A1a (in D), that binding to A1-1 (which contains A1a) is cooperative, and is eliminated by mut7, which has altered only the 4 nt within A1a shown in B (“M7”).
F: binding to large, overlapping probes A0–A5 (as indicated at the top) by a combination of all 3 proteins (lanes marked “+” at the top). The position of the main cooperative complex is indicated by a filled arrowhead. Note that binding to A1 is by far the strongest, but that there is also detectable binding to A2 (which contains A2a and A2-1, shown in E), A5, and A0.
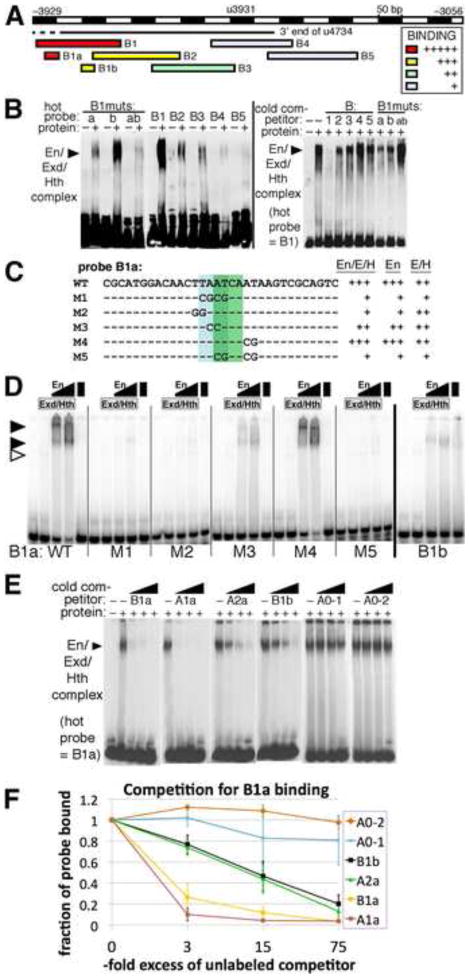
Fig. 6. Cooperative binding by Engrailed. Extradenticle and Homothorax to En-responsive region B within CRM u4734.
A: summary of in vitro binding to probes from region B by En+Exd+Hth. Relative strength of binding in EMSAs is indicated by colors, listed in the inset. Examples are shown in B.
B: Left panel: binding to large labeled (“hot”) probes in region B by En+Exd+Hth (lanes labeled with “+”). The labeled probes used are indicated across the top: under B1muts, “a” has the M1 mutation within B1a (shown in C), “b” has a mutation within B1b that destroys cooperative binding (data not shown), and “ab” has both. The position of the main cooperative complex is indicated by a filled arrowhead. Right panel: competition for binding to B1 by BIBS and by mutant versions of B1, as indicated across the top. Note that B1 competes very effectively, while other oligonucleotides are less effective. Under B1muts, B1 carrying mutations “a”, “b”, or both (“ab”) was used as unlabeled (“cold”) competitor.
C: summary of in vitro binding to probe B1a and mutated versions. Indicated to the right of each probe sequence is the observed strength of complex formation by En+Exd+Hth (“En/E/H”), En alone, or Exd+Hth (“E/H”) relative to that for B1a (“WT”) as determined by EMSAs, some of which are shown in D.
D: binding to probe B1a and mutant versions (indicated at the bottom), and to B1b, by Exd+Hth, En and a combination of all 3, at 2 different concentrations of En (as indicated at the top, see Materials and Methods). The positions of cooperative complexes formed with all 3 proteins are indicated by filled arrowheads, and the position of the Exd+Hth complex by an open arrowhead. Note that binding to B1a by all 3 proteins is cooperative, that there is virtually no cooperative complex with either mutant M1, M2, or M5, and that binding to B1b appears weaker and less cooperative than binding to B1a.
E: competition for binding by En+Exd+Hth to B1a by unlabeled oligonucleotides representing the sites listed across the top, at about 3, 15, and 75-fold molar excess over probe (B1a) from left to right in each case. Oligo sequences used were: B1a: CGCATGGACAACTTAATCAATAAGTCGCAGTC, A1a: GCAAACAACTAATTAAGTCATCAGGATGAAAG, A2a: ATGAGGTTCAAAATGAGCCATCACCCATTGAC, B2a: GGTGTGGAACTGATCAATTATAGTGGGTGTTG, A0-1: ACACTTGAAAAATATTCATCAGTGAATAAGac, A0-2: tTCTTTTTCCGTTGACCATCAGCTGCTGTCCa. Note that both A1a and B1a compete very well, while A2a and B1b compete somewhat well, and A0-1 and A0-2 compete relatively poorly.
F: Quantitation of competition binding assays. Three trials of the competition assays shown in E were quantified using densitometry and Image J software. The averages and standard deviations of the fraction of probe bound at each concentration of competitor are plotted, for the oligonucleotides listed in the inset. Note that A1a competes best, indicating that it has the highest affinity for the combination of En+Exd+Hth at the concentrations used, B1a competes only slightly less well, while A2a and B1b each show an ability to compete that indicates an intermediate affinity, and A0-1 and A0-2 show little ability to compete at these concentrations.
The non-radioactive electromobility shift assay (EMSA) was used to survey i1523 and the 3′ end of u4734 for binding by En/Exd/Hth to DNA fragments of 150–200 bp (A0–A5 and B1–B5). Probes were prepared using two-step PCR. Each fragment was amplified with specific primers (given in Fig. S1) that contained the same sequence tag (CGCTACGACTCACTATAGGGC) at their 5′ end. The purified PCR products was used as template in a second PCR, using a biotinylated-5′-primer against the tag. EMSA reactions were analyzed on mini-gels (Thermo scientific), and transferred to MagnaGraph membranes (GE). Probes were visualized by incubating with SA-HRP, followed by the ECL reaction (GE). “Cold” competitors were single PCR products.
Results
Multiple enhancers are regulated by Eve and En to delimit stripe expression at different stages of embryogenesis
Eve limits the expression of each slp stripe within the early 7-stripe pattern (Fujioka et al., 1995; Jaynes and Fujioka, 2004), while En is required to restrict the posterior border of each slp stripe within the 14-stripe pattern (Cadigan et al., 1994). Previous analysis suggested that slp is a directly repressed target of En (Jaynes and Fujioka, 2004; Kobayashi et al., 2003). As a test of whether these related repressors act through the same or separate binding sites, we conducted an analysis of minimal stripe CRMs (Fujioka and Jaynes, 2012) using transient ectopic expression from an inducible heat shock (hs) promoter to determine their responses to Eve and En. The results of this analysis are summarized in Fig. 1, and described below.
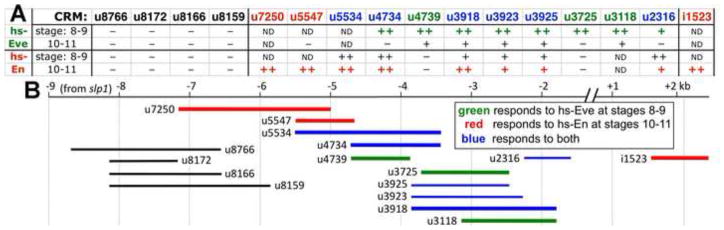
Fig. 1. Responsiveness of slp CRMs to ectopic Eve and En.
A: 1st row: transgenic lines carrying various CRMs. 2nd and 3rd rows: response to ubiquitous expression of Eve from an inducible heat shock promoter at stages 8–9 and 10–11, respectively. 4th and 5th rows: response to ubiquitous expression of En at stages 8–9 and 10–11, respectively. “++”, “+” and “−” indicate the degree of repression: strong, moderate, and none, respectively. ND: not done.
B: map of slp stripe CRMs color coded with their responsiveness to ectopic Eve and En, as indicated in the inset box.
To display the location of each CRM within the locus, each of our CRM names begin with a letter, indicating whether it is upstream of slp1 (u), or within the slp1–slp2 intervening region (i). This letter is followed by 4 digits, the first two corresponding to the 5′ CRM end point and the last 2 to the 3′ end point, in hundreds of kb (see Fig. 2 for a map of the slp locus). For example, u4734 extends from −4.7 to −3.4 kb upstream of the slp1 TSS, while i1523 extends from +1.5 to +2.3 kb downstream of the slp1 TSS (see Fig. S1 for exact locations of all CRMs and probes used in this paper).
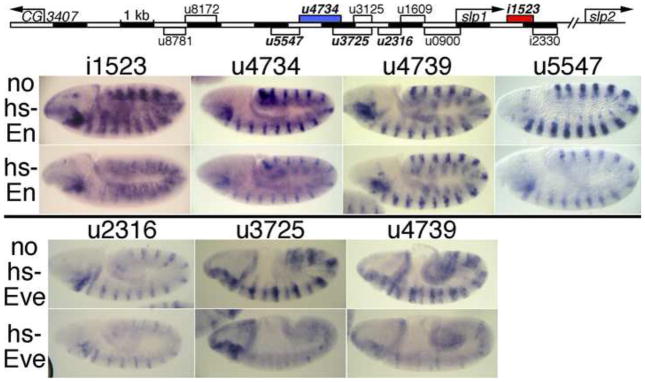
Fig. 2. Repression of CRM activity bv ectopic En and Eve.
Map at top: CRMs named in bold italics drive a 14-stripe slp1-like pattern (Fujioka and Jaynes, 2012), adjacent to En stripes. The red and blue filled boxes indicate CRMs where we identified functional En binding sites, as shown in later figures.
All rows: embryos carried the transgenic CRM-lacZ reporter indicated across the top.
2nd and 4th rows: embryos also carried a heat shock-inducible transgene driving ubiquitous En (2nd row) or Eve (4th row) expression. Embryos were heat shocked and stained as described in Materials and Methods. Note in the 2nd row that expression driven by each CRM except u4739 is significantly repressed by En, and that all of the CRMs in the 4th row are repressed by Eve, u2316 less completely so than the others, as indicated in Fig. 1.
Overall, we identified 4 non-overlapping regions that are regulated by En, and 3 by Eve. CRM i1523, which drives 14 stripes at stages 9–10 (Fig. 2, (Fujioka and Jaynes, 2012)), was rapidly and efficiently repressed by hs-En (Figs. 1, 2), consistent with direct regulation by En. We chose this CRM for analysis of DNA binding by En and its cofactors (see below). Similarly, CRM u2316 drives 14 stripes at stages 9–11 (Fujioka and Jaynes, 2012). At these stages, the 7 strong Eve stripes are fading away (Frasch et al., 1987; Macdonald et al., 1986), and both slp and en are required to maintain the parasegment border (Cadigan et al., 1994; Kobayashi et al., 2003), which is between the posterior edge of each slp stripe and the anterior edge of each En stripe. Not unexpectedly, hs-En expression repressed this enhancer at all stages (Fig. 1), while hs-Eve had only a weak effect when expression is first initiated (Fujioka and Jaynes, 2012) (stage 9, when hs-En had a strong effect), and no significant effect at stages 10–11 (Fig. 1). This suggests that Eve and En may act through distinct sites (confirmed by results below), and/or that they may require different cofactors that are differentially expressed.
CRMs u3925 and u3725 each drive expression at stages 5 through 10, including both a 7- and a 14-stripe pattern (Fujioka and Jaynes, 2012), suggesting regulation by both Eve and En. Consistent with direct regulation by both, hs-Eve efficiently repressed u3925 at stages 7–9, while hs-En did so at stages 8–10 (Fig. 1). While u3725 responded to hs-Eve similarly to u3925, it was not repressed effectively by hs-En, indicating En-responsive sites between −3.9 and −3.7 kb. This 200 bp region is also part of CRM u4734, and turns out to contain an essential En binding site (see below). Consistent with truncated forms of this CRM responding to Eve but not En, u3125 is expressed only in a 7-stripe pattern (Fujioka and Jaynes, 2012; Prazak et al., 2010), and its expression fades at about the same time that Eve expression fades (data not shown).
Similar to u3925, CRM u4734 is active at stages 5 through 11, including both a 7- and a 14-stripe pattern. While induction of hs-En repressed u4734 at all stages (Figs. 1, 2), hs-Eve repressed it at stages 7–8, but not stage 10 (Fig. 1). When u4734 was truncated from its 3′ end to generate u4739, responsiveness to hs-En was lost (Figs. 1, 2), but not to hs-Eve. This loss of En responsiveness is similar to that observed when part of the same region was deleted from u3725 (above), and confirms that this region is likely to contain En binding sites. Notably, the response to hs-Eve at stages 7–9 was not affected by truncating either u3925 or u4734 (Fig. 1), suggesting that Eve and En utilize distinct binding sites, even within these commonly regulated CRMs.
CRM u5547 reinforces the same theme. It drives 14 stripes beginning at stage 9, as Eve expression is fading. Consistent with this, hs-En, but not hs-Eve, repressed it efficiently (Figs. 1, 2). Again, En and Eve appear to be acting through largely if not entirely distinct regions.
CRM u8172 and those tested regions that contain it (u8766, u8166, u8159, Fig. 1) drive an aberrant pattern at stages 6 through 9 that includes some cells within the eve domain that do not express slp (Fujioka and Jaynes, 2012; Prazak et al., 2010). Consistent with this ectopic expression, neither hs-Eve nor hs-En repressed it efficiently (Fig. 1). CRMs which extend beyond the core u8172 region by more than 500 bp upstream and more than 1.4 kb downstream still drive ectopic expression and do not respond well to hs-Eve. This confirms the suggestion of a recent study that sequences downstream of this region are normally responsible for inhibiting the activity of u8172 within the eve domain (Prazak et al., 2010).
A slp intergenic regulatory module contains En/Exd/Hth binding sites
Based on responsiveness to hs-En, we selected i1523 for analysis of binding in vitro. A genome-wide survey of En binding regions (Celniker et al., 2009; Negre et al., 2011) identified this CRM as the single high-probability peak within the slp stripe-forming region. Within i1523 are found adjacent sequences that closely match consensus sites for both En/Hox and a complex of Exd and Hth (Exd/Hth). The immediately surrounding region is well conserved through drosophilid evolution (Fig. 3A). We performed electromobility shift assays (EMSAs) using a 33 bp oligonucleotide centered on the consensus sites (A1a, Fig. 3A–D). This probe was bound weakly by either En or Exd/Hth, but was bound strongly by En with Exd/Hth (Fig. 3B,C,D, A1a). We tested the importance for binding of the two consensus sequences within A1a. Mutating the En consensus abolished En binding and cooperative binding with Exd/Hth, but not Exd/Hth binding (Fig. 3B, M6, and Fig. 3D, A1a-mut6). Mutating both consensus sites caused a complete loss of binding (Fig. 3B, M7, and Fig. 3D, A1a-mut7). Analysis of other mutant sites suggested that both consensus sequences contribute to En binding in the absence of Exd/Hth, while Exd/Hth binding in the absence of En mainly depends on the Exd/Hth consensus region (Fig. 3B and data not shown). Similar cooperative binding that depended on the consensus sites was seen using larger probes centered on A1a (Fig. 3C, A1a′ and A1-1; Fig. 3E, A1-1 and A1-1-mut7).
We used this cooperative binding assay with a series of 6 larger, overlapping probes of 150–200 bp (A0 – A5) to survey the entire i1523 CRM. Only A1, centered on A1a, showed strong binding, while 3 others (A0, A2, and A5) gave signals significantly above background (Fig. 3C, F). Within A2, we tested a 25 bp sequence (A2a, Fig. 3C, E) centered on an Exd core consensus site (ATCA), which also contained a Hth core consensus (TGAC), for cooperative binding by En and Exd/Hth. We found that although binding was considerably weaker than for A1a, it was nonetheless strongly cooperative (Fig. 3E, left panel). In contrast, a probe that corresponds to the half of A2 without A2a (A2-1, Fig. 3C) showed only weak binding by Exd/Hth, and no cooperativity with En (Fig. 3E, right panel, A2-1), suggesting that most of the cooperative binding activity of A2 is due to A2a. While the A1a region shows clear conservation among 12 species of Drosophila, A2a is conserved only among the more closely related species (analyzed by Evoprinter, not shown) (Clark et al., 2007; Odenwald et al., 2005).
As a test of whether cooperative binding sites could be localized within a weak binding region such as A0, we used a series of small probes to subdivide A0 (Fig. 3C, A0-1 – A0-5). We found only weak cooperative binding to the A0-1 region (Fig. 3C and data not shown), which contains the Exd/Hth core consensus (green boxed in Fig. 3A, B) but no canonical En binding core (ATTA; see Fig. S1 for more detailed sequence information).
Overall, within i1523, there is one very strong, cooperative binding site for En/Exd/Hth (A1a), at least one weaker but still cooperative site (A2a), and other quite weak binding sites. We next used transgenic analysis to test whether these sites are functionally significant.
The high affinity, cooperative binding site within i1523 is redundant with a second, lower affinity, highly cooperative site
The i1523 CRM drives reporter gene (lacZ, which encodes β–galactosidase, or βgal) expression in 14 stripes that are wider than endogenous slp stripes, but still mostly excluded from the En domain. There is some overlap with En in dorsal regions, mostly in parasegments 9–12 (Fig. 4, left column), suggesting that although this CRM is efficiently repressed by hs-En (Figs. 1, 2), it is nonetheless less stringently repressed by En than the endogenous gene. In addition, the width of the stripes indicates that it is missing sites that normally prevent expression anterior to endogenous slp within each parasegment (possibly sites for Odd-skipped binding, based on its expression pattern).
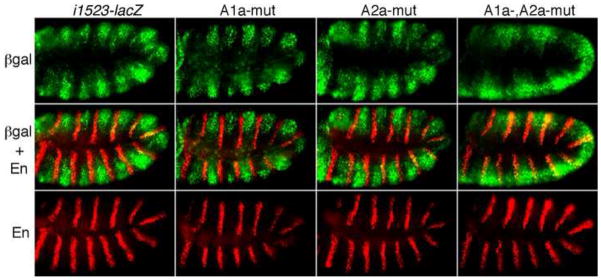
Fig. 4. Redundant function of En/Exd/Hth binding sites within i1523.
Reporter transgenes with CRM i1523, and En/Exd/Hth site-mutated forms, driving lacZ are indicated across the top. βgal expression is visualized in green, and En in red, using appropriate antibodies. Note that βgal expression driven by i1523 is mostly non-overlapping with En stripes, although there is some overlap in dorsal regions, and that βgal overlaps extensively with En when both binding sites are mutated (A1a-, A2a-mut), but not when either one alone is mutated (A1a-mut, A2a-mut).
Our in vitro analysis (above) indicated that a pair of 2-nt changes was sufficient to abolish binding by En, as well as cooperative binding by En with Exd/Hth, to the single strong site within i1523 (Fig. 3B, M7, and Fig. 3D, A1a-mut7). We tested whether this alteration compromises repression in vivo by incorporating it into the i1523-lacZ transgene. Surprisingly, given that A1a is the only strong, cooperative site within this CRM and the fact that the response of i1523 to En is already weaker than that of endogenous slp, we did not observe significant derepression relative to unmutated i1523-lacZ (Fig. 4, 2nd column, A1a-mut, vs. 1st column). Similarly, introducing a mutation that abolishes En/Exd/Hth binding in vitro to the weaker A2a site (data not shown) caused no significant derepression (Fig. 4, 3rd column, A2a-mut). However, when both mutations were introduced simultaneously into the i1523-lacZ transgene, expression was strongly derepressed in the En domain, particularly in ventral regions of the embryo (Fig. 4, A1a-,A2a-mut). Thus, despite the apparent difference in their affinity in vitro, there is redundancy among the En/Exd/Hth cooperative binding sites in i1523 for preventing slp expression within the En domain.
The En repression module within CRM u4734 contains an essential En/Exd/Hth binding site
We next focused on CRM u4734, which drives strong expression that overlaps in space and time with the weaker expression driven by i1523 (Fujioka and Jaynes, 2012). Transgenic dissection suggested that u4734 contains an essential En-responsive element between −3.9 and −3.4 kb (relative to the slp1 TSS). In addition to a loss of hs-En responsiveness when this region was deleted to generate u4739, u4739 also drives wider stripes than does u4734 (Figs. 1, 2). We tested whether these wider stripes extend into the en domain by double-staining for lacZ and en RNA, and found that while u4734-lacZ is not expressed in the en domain, u4739-lacZ expression overlaps en stripes at embryonic stages 10–11, most strongly in ventral regions (Fig. 5). As described above, response to hs-En by u3925 was lost when the region −3.9 to −3.7 kb was deleted to generate u3725 (Fig. 1), suggesting that the En response of u4734 might be due to this region. We identified closely situated matches to core consensus binding sites for both En and Exd at two places within this region. We therefore conducted an in vitro analysis of binding by these proteins to the region between −3.9 and −3.4 kb.
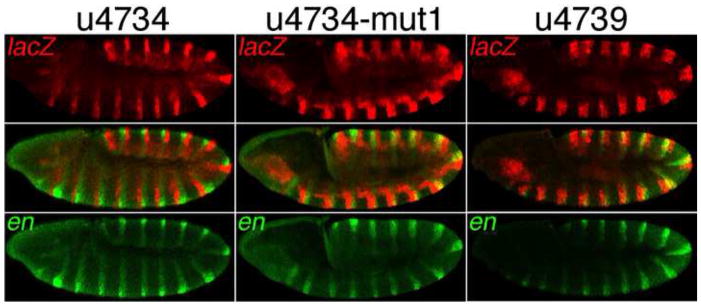
Fig. 5. An essential conserved En/Exd/Hth consensus binding site in u4734.
Reporter transgenes were driven by the CRMs indicated across the top. lacZ mRNA was visualized in red and en mRNA in green using FISH. Note that u4734-mut1, which includes a mutation (M1) within the cooperative En/Exd/Hth binding site B1a (see Fig. 6), as well as u4739, drive extensive expression within the en stripes, while u4734 does not.
We used overlapping 150–200 bp probes to survey the entire region for binding by En/Exd/Hth (Fig. 6A, B). We saw very strong binding to probe B1, which contains both of the regions (B1a and B1b, for sequence see Fig. 7 and Fig. S1) that have closely situated matches to En and Exd consensus sequences. In addition, we saw considerably weaker binding to probes B2 and B3 (Fig. 6A, B). We then used competition assays to confirm the relative binding strengths of these probes. Cold B1 competed much better for labeled B1 than did either cold B2 or B3, and competition by B4 and B5 was minimal. We introduced mutations that changed both the En and Exd consensus sequences within B1a, and separately within B1b, and tested the effects in vitro in two ways. First, we labeled the resulting B1 mutant probes and tested their ability to support cooperative binding, and second, we tested their ability to compete with unmutated, labeled B1 probe for binding. We found that mutating either pair of En-Exd consensus sites reduced binding by En/Exd/Hth, while mutating both pairs almost abolished both direct binding (Fig. 6B, left panel) and the ability to compete with labeled B1 for binding (right panel). The B1a region is very well conserved among Drosophila species, while the B1b region is not well conserved. We analyzed B1a directly for binding in vitro, and found that it supported strong, cooperative binding by En with Exd/Hth (Fig. 6C, D). We also tested binding to B1b in vitro, and found that although it bound En alone about as well as did B1a, cooperativity with Exd/Hth was considerably weaker than for either B1a or for the two sites identified within i1523, A1a or A2a (Fig. 6A and data not shown). That cooperativity on B1b is weaker than on A2a is based on the fact that B1b binding by En alone or by Exd/Hth alone was greater than A2a binding. Despite this, binding of the cooperative complex by the two sites is similar. This can be explained by a greater cooperativity of En with Exd/Hth on A2a.
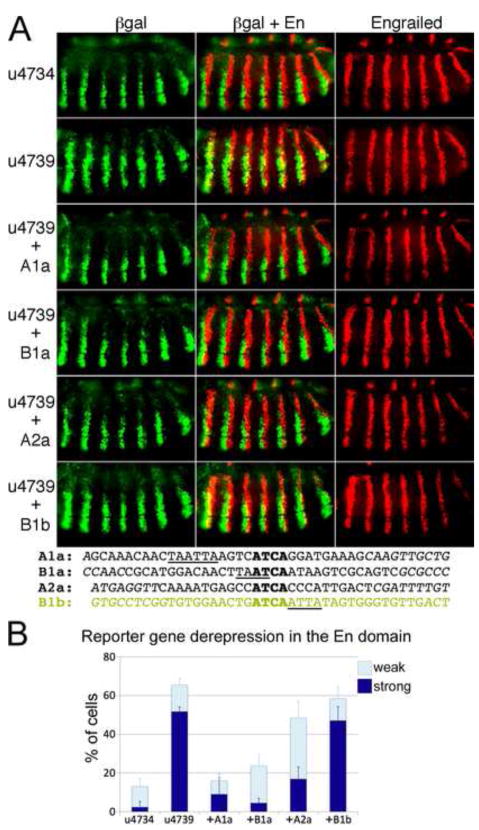
Fig. 7. Conserved cooperative En/Exd/Hth binding sites act as repression elements in embryos.
A: Reporter transgenes were driven by u4734 and derivatives, as indicated on the left. βgal expression was visualized in green, and En in red, using appropriate antibodies. 1st row, u4734; 2nd row, u4739; 3rd–6th rows: 40 – 50 bp conserved sequence blocks containing the En/Exd/Hth binding sites A1a, B1a, A2a, and B1b, respectively, attached to the 3′ end of u4739 (sequences given at the bottom; see also Fig. S1). Note that most of the derepression of βgal in the En domain with u4739 was reversed by addition of B1a, A1a, and A2a, but not B1b, which remains similar to u4739. Bottom: sequences added to u4739 in each case, with Exd core consensus sequences aligned (bold), and En core consensus sequences underlined.
B: Quantitation of repression by binding sites in A. Reporter gene expression in individual cells within En stripes was assessed as described in Materials and Methods and illustrated in Fig. S2. Three levels of expression were distinguished: none detected, weak expression (clearly below that seen within endogenous slp stripes), and strong expression (comparable to that within slp stripes). The percentage of cells showing either strong or weak expression was averaged over at least 4 microscopic fields (at least 450 cells total, from at least 4 different embryos), and this average is graphed, along with the corresponding standard deviation.
We next identified clustered point mutations within the consensus sequences of B1a that virtually eliminated binding (Fig. 6C, D), and introduced one of these (M1, Fig. 6C) into the u4734-lacZ transgene for analysis in vivo. We found that mutating B1a causes derepression in the En domain that is indistinguishable from the effect of deleting the entire region from −3.9 to −3.4 kb (Fig. 5, u4734-mut1 vs. u4739). Thus, this conserved, cooperative binding site is essential for En-dependent repression of u4734, despite the existence of another set of nearby consensus binding sites (in B1b, Figs. 6A, 7).
We directly compared the affinities of the 4 En/Exd/Hth binding sites that we localized within i1523 and u4734 using competition assays. We quantified the amounts of B1a, B1b, A1a and A2a required to compete with labeled B1a for binding by En/Exd/Hth (Fig. 6E, F). The results show that A1a has a comparable, slightly higher affinity than does B1a, while A2a and B1b have lower affinities that are very similar to each other. All of these relative affinities, including those of the negative controls A0-1 and A0-2, are entirely consistent with the apparent relative affinities from the direct binding assays (Figs. 3C–E, 6A, D). Thus, A1a (functionally important but redundant with A2a) and B1a (essential) have clearly higher affinities than either A2a (functionally important but redundant with A1a) or B1b (nonessential), which are very similar to each other.
The highly cooperative En/Exd/Hth binding sites B1a, A1a, and A2a are each sufficient for repression within the En domain
Deletion of the En repression module from CRM u4734 to generate u4739 causes strong derepression in the En stripes (Fig. 5). We asked whether the identified En/Exd/Hth binding sites are sufficient to substitute for this repression module. When a 40–50 bp conserved block of genomic sequence (see Fig. S1) containing either of the highest affinity sites, A1a or B1a, was added to the 3′ end of u4739 in the context of the u4739-lacZ transgene, βgal expression was excluded from the En stripes. Slight derepression was observed in a few cells relative to u4734, which contains the entire repression module (Fig. 7A, top 4 rows, and Fig. 7B). The lower affinity but highly cooperative site A2a was slightly less effective, with significantly more cells showing weak derepression (Fig. 7A, B). In contrast, the lower affinity and less cooperative site B1b (in a 40 bp natural context) was not sufficient to confer repression in the En domain, but was derepressed almost as completely as u4739 (Fig. 7A, B).
Discussion
slp CRMs contain distinct Eve- and En-responsive regions
Consistent with the fact that Eve is expressed earlier than En, with some overlap at embryonic stages 8–9, slp CRMs tended to respond to ectopically expressed Eve at earlier stages than to En (Fig. 1). Our transgenic dissections further showed that they have distinct responsive regions within CRMs, suggesting that many of their binding sites are distinct (Fig. 1 and 2). This is somewhat surprising because they are both homeodomain-containing repressors that set the posterior borders of slp stripes, and they have been seen to have similar in vitro binding specificities (Hoey and Levine, 1988). A possible explanation is that they cooperate in DNA binding with different cofactors, making their functional sites distinct. Despite detailed analyses of Eve function in segmentation, no candidate co-factors for specifying target genes have emerged.
A recent study showed that CRM u8172 drives ectopic expression within odd-numbered parasegments in cells that normally do not express detectable levels of slp RNA. However, when combined with the promoter-proximal CRM u3125, which drives properly restricted expression within even-numbered parasegments, ectopic expression is repressed, suggesting that an Eve-responsive element resides within this region (Prazak et al., 2010). Consistent with these findings, transgenes containing this region (u3925, u3725, Fig. 1) responded to ectopically expressed Eve (Fig. 1), and rescue-type transgenes carrying u8172 without this region drove ectopic Slp, causing embryonic defects (Fujioka and Jaynes, 2012).
Cooperativity and functional redundancy
Recently, we found that a striking number of distinct CRMs surrounding the slp1 transcription unit drive expression that overlaps in both space and time. Extensive dissection of this regulatory region and rescue of slp mutants with various transgenes suggested that apparent redundancy may be necessary to provide fully functional levels of expression across the various stages of slp expression. Here, we have shown that there are functionally redundant En/Exd/Hth binding sites within CRM u1523. In vitro binding analysis identified a strongly cooperative binding site (A1a, Figs. 3, 4) and a weaker, but still highly cooperative site (A2a, Figs. 3, 4). Despite the apparent difference in in vitro binding affinity, either site is sufficient to confer repression in the En domain (Fig. 7), and both sites must be mutated to cause significant derepression (Fig. 4). Thus, apparent redundancy exists at multiple levels in slp regulation. Whether apparent redundancy at this level has a function in increasing the robustness of functional gene expression within the organism, as does apparent redundancy among multiple enhancers regulating the same gene (Frankel et al., 2010; Perry et al., 2010), remains to be determined. Furthermore, cooperativity with cofactors in vitro seems to be a significant indicator of function in vivo, in addition to affinity. We found that while the B1b site has the same apparent affinity as A2a (Fig. 6F), A2a confers considerably stronger repression activity (Fig. 7), and shows greater cooperativity in binding by En with Exd/Hth (Figs. 3E and 6F, described in Results). The discrepancy between relative affinity and functionality may be attributed to the challenge of reproducing functional binding conditions in vitro, where protein-protein interactions leading to cooperativity may be less sensitive to the differences in conditions than are protein-DNA interactions. Relatedly, competition with a variety of DNA binding proteins in vivo for sites on the DNA may lead to a greater reliance on cooperativity in vivo for occupancy of functional sites.
Previous studies indicated that En requires the Hox co-factors Exd and Hth to efficiently repress slp, especially in the anterior half of the embryo (Alexandre and Vincent, 2003; Kobayashi et al., 2003), and En was found to act cooperatively on target sites in the distalless gene with both Exd/Hth and posteriorly-expressed Hox gene products (Gebelein et al., 2004; Lelli et al., 2011). Although it remains possible that the relatively weak, yet functional binding site (A2a) within i1523 might bind En with other cofactors in addition to Exd/Hth, our dissection and construction experiments with this and other sites have not revealed any clear anterior-posterior differences in their activity that might suggest a functional interaction with cofactors such as Hox proteins that are restricted in expression along the anterior-posterior axis. Nonetheless, previous studies suggested that regulation of slp by En might utilize posterior-specific factors (Alexandre and Vincent, 2003; Kobayashi et al., 2003). Further analysis will be required to more fully explore this possibility.
The relative arrangement of consensus En and Exd sites that facilitate cooperative binding appears to be quite flexible. For example, the A2a site contains no canonical consensus core for En binding (ATTA), while for the other two functional sites, the distance between the centers of the En and Exd sites is 10–12 bp for A1a (Figs. 3, 7) and only 2 bp for B1a (Fig. 7). The latter is reminiscent of En-Exd/Hth binding in distalless (Gebelein et al., 2004), where simultaneous Hox binding occurs, although the position of the En site is on the opposite side of the Exd core consensus ATCA. This relative arrangement of En and Exd sites (En binding 5′ of the Exd core ATCA) is seen for all of the functional sites analyzed here (Figs. 3, 6, and data not shown from a limited mutational analysis of A2a). This arrangement is similar to the relative positions of Hox and Exd binding to sites where there is no En involvement (Slattery et al., 2011; Uhl et al., 2010). The flexibility overall is consistent with that seen for Exd/Hth binding in conjunction with the Hox gene products (Uhl et al., 2010), and suggests that while homeodomain family transcription factors are able to function combinatorially in vivo on a wide variety of binding sites, there are significant constraints on the positions of contact by the individual homeodomains. A full understanding of the similarities and differences between En binding in conjunction with Exd and Hth, and Hox binding with these cofactors, will require further investigation.
We found that the highly cooperative, strong En/Exd/Hth binding site B1a was both necessary and sufficient for repression of u4734 in the En domain. However, it did not fully substitute for the entire repression element that contains it, located between −3.9 and −3.4 kb from the slp1 TSS (Fig. 7). This finding suggests that there may be other functional En binding sites in this region. Consistent with this, in vitro binding suggested that other subregions (B2 and/or B3, Fig. 6A) harbor some binding activity. Thus, like i1523, there may be partial redundancy in En complex binding within u4734, despite the existence of a single essential binding site.
Conservation of functional binding sites
We have established the functional significance of three cooperative En/Exd/Hth binding sites within slp. Interestingly, two of them are well conserved among the 12 species of Drosophila whose genomes have been sequenced (Clark et al., 2007), and the other site is conserved within the more closely related species. The duplication that generated the twin slp transcription units apparently took place before the divergence of these 12 species, as all drosophilids (but not mosquitoes) contain two tandem slp-related protein coding regions (Fujioka and Jaynes, 2012). This might suggest that the two conserved En/Exd/Hth sites were duplicated along with the locus as a whole. It has been shown that Drosophila enhancers contain clusters of conserved sequences blocks (Brody et al., 2008; Kuzin et al., 2009), and the two CRMs analyzed in this study contain such conserved sequence clusters. However, the patterns of conservation in the regions surrounding the conserved En/Exd/Hth sites do not suggest that they are directly related to each other. Furthermore, both CRMs are more closely linked to slp1 than to slp2. Clearly, there have been other chromosomal rearrangements in the history of the slp locus, precluding a simple description of its evolution.
A recent study investigating the genome-wide distribution of En binding showed a peak on i1523, but not on u4734 (Celniker et al., 2009; Negre et al., 2011). The data were derived from 7–24 hr-old embryos, which were mostly at later stages than those at which these CRMs are active. In addition, the data show peaks where our analysis has not identified functional CRMs. Such sites may function to assist those within the core enhancer regions, or they might be functional during larval or adult stages to keep slp in the off state. Alternatively, they might not be functionally important. Further study will be required to address these issues.
Supplementary Material
Highlights.
multiple slp CRMs are targets of repression by both Engrailed and Even-skipped
despite similar individual binding specificities, they have distinct target sites
Engrailed binds with the Hox cofactors Exd and Hth to sites in two separate CRMs
several essential target sites were localized and their functionalities compared
affinity in vitro, by itself, poorly predicts functionality in vivo
Acknowledgments
We thank Guizhi Sun and Galina Yusibova for excellent technical assistance, and the Developmental Studies Hybridoma Bank (supported by NICHD and maintained by the Univ. of Iowa Dept. of Biology) for anti-En (4D9). Confocal imaging and DNA sequencing were carried out in Kimmel Cancer Center facilities, which are supported in part by NCI Cancer Center Grant P30CA56036. This work was supported by the National Institutes of Health (F31-GM083648 to ZCC, GM079428 to BG and GM050231 to JBJ) and the National Science Foundation (MCB-0818118 to JBJ and MF).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Alexandre C, Vincent JP. Requirements for transcriptional repression and activation by Engrailed in Drosophila embryos. Development. 2003;130:729–739. doi: 10.1242/dev.00286. [DOI] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody T, Rasband W, Baler K, Kuzin A, Kundu M, Odenwald WF. Sequence conservation and combinatorial complexity of Drosophila neural precursor cell enhancers. BMC Genomics. 2008;9:371. doi: 10.1186/1471-2164-9-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Grossniklaus U, Gehring WJ. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994;8:899–913. doi: 10.1101/gad.8.8.899. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Dillon LAL, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, Micklem G, Piano F, Snyder M, Stein L, White KP, Waterston RH, mod EC. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe CP, Brown SJ. Evolutionary flexibility of pair-rule patterning revealed by functional analysis of secondary pair-rule genes, paired and sloppy-paired in the short-germ insect, Tribolium castaneum. Dev Biol. 2007;302:281–294. doi: 10.1016/j.ydbio.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Eisen MB, Smith DR, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart W, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chappie C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia ACL, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li R, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long M, Low L, Lozovsky E, Lu J, Luo M, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MAF, O’Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VLS, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GKS, Wu C-L, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang P, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D’Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu X, Liu J, Liu S, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O’Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong D, Zimmer A, Zwirko Z, Alvarez P, Brockman W, Butler J, Chin C, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Datta RR, Small S. Gene regulation: piecing together the puzzle of enhancer evolution. Curr Biol. 2011;21:R542–543. doi: 10.1016/j.cub.2011.06.026. [DOI] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature. 2010;466:490–493. doi: 10.1038/nature09158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M, Warrior R, Tugwood J, Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes Dev. 1988;2:1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB. Regulation of a duplicated locus: Drosophila sloppy paired is replete with functionally overlapping enhancers. Dev Biol. 2012;362:309–319. doi: 10.1016/j.ydbio.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Jaynes JB, Goto T. Early even-skipped stripes act as morphogenetic gradients at the single cell level to establish engrailed expression. Development. 1995;121:4371–4382. doi: 10.1242/dev.121.12.4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Yusibova GL, Patel NH, Brown SJ, Jaynes JB. The repressor activity of Even-skipped is highly conserved, and is sufficient to activate engrailed and to regulate both the spacing and stability of parasegment boundaries. Development. 2002;129:4411–4421. doi: 10.1242/dev.129.19.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Yusibova GL, Zhou J, Jaynes JB. The DNA-binding Polycomb-group protein Pleiohomeotic maintains both active and repressed transcriptional states through a single site. Development. 2008;135:4131–4139. doi: 10.1242/dev.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebelein B, Culi J, Ryoo HD, Zhang W, Mann RS. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell. 2002;3:487–498. doi: 10.1016/s1534-5807(02)00257-5. [DOI] [PubMed] [Google Scholar]
- Gebelein B, McKay DJ, Mann RS. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature. 2004;431:653–659. doi: 10.1038/nature02946. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Pearson RK, Gehring WJ. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992;6:1030–1051. doi: 10.1101/gad.6.6.1030. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemskerk J, DiNardo S, Kostriken R, O’Farrell PH. Multiple modes of engrailed regulation in the progression towards cell fate determination. Nature. 1991;352:404–410. doi: 10.1038/352404a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoey T, Levine M. Divergent homeo box proteins recognize similar DNA sequences in Drosophila. Nature. 1988;332:858–861. doi: 10.1038/332858a0. [DOI] [PubMed] [Google Scholar]
- Ingham PW, Gergen JP. Interactions between the pair-rule genes runt, hairy, even-skipped and fushi tarazu and the establishment of periodic pattern in the Drosophila embryo. Development Suppl. 1988;104:51–60. [Google Scholar]
- Jaynes JB, Fujioka M. Drawing lines in the sand: even skipped et al and parasegment boundaries.[erratum appears in Dev Biol 2004 Aug 1;272(1):277–8] Dev Biol. 2004;269:609–622. doi: 10.1016/j.ydbio.2004.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Fujioka M, Tolkunova EN, Deka D, Abu-Shaar M, Mann RS, Jaynes JB. Engrailed cooperates with extradenticle and homothorax to repress target genes in Drosophila. Development. 2003;130:741–751. doi: 10.1242/dev.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. doi: 10.1126/science.1099247. [DOI] [PubMed] [Google Scholar]
- Kuzin A, Kundu M, Ekatomatis A, Brody T, Odenwald WF. Conserved sequence block clustering and flanking inter-cluster flexibility delineate enhancers that regulate nerfin-1 expression during Drosophila CNS development. Gene Exp Patterns. 2009;9:65–72. doi: 10.1016/j.gep.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli KM, Noro B, Mann RS. Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity. Proc Natl Acad Sci USA. 2011;108:21122–21127. doi: 10.1073/pnas.1114118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PM, Ingham P, Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986;47:721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoukian AS, Krause HM. Control of segmental asymmetry in Drosophila embryos. Development. 1993;118:785–796. doi: 10.1242/dev.118.3.785. [DOI] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, Li Z, Ishii H, Spokony RF, Chen J, Hwang L, Cheng C, Auburn RP, Davis MB, Domanus M, Shah PK, Morrison CA, Zieba J, Suchy S, Senderowicz L, Victorsen A, Bild NA, Grundstad AJ, Hanley D, MacAlpine DM, Mannervik M, Venken K, Bellen H, White R, Gerstein M, Russell S, Grossman RL, Ren B, Posakony JW, Kellis M, White KP. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenwald WF, Rasband W, Kuzin A, Brody T. EVOPRINTER, a multigenomic comparative tool for rapid identification of functionally important DNA. Proc Natl Acad Sci USA. 2005;102:14700–14705. doi: 10.1073/pnas.0506915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990;4:1209–1223. doi: 10.1101/gad.4.7.1209. [DOI] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prazak L, Fujioka M, Gergen JP. Non-additive interactions involving two distinct elements mediate sloppy-paired regulation by pair-rule transcription factors. Dev Biol. 2010;344:1048–1059. doi: 10.1016/j.ydbio.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V, Irion U, Wilson R, Grosskortenhaus R, Leptin M. Control of cell fates and segmentation in the Drosophila mesoderm. Development. 1997;124:2915–2922. doi: 10.1242/dev.124.15.2915. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, Mann RS. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. Near-reciprocal phenotypes caused by inactivation or indiscriminate expression of the Drosophila segmentation gene ftz. Nature. 1985;318:677–680. doi: 10.1038/318677a0. [DOI] [PubMed] [Google Scholar]
- Tolkunova EN, Fujioka M, Kobayashi M, Deka D, Jaynes JB. Two distinct types of repression domain in engrailed: one interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol Cell Biol. 1998;18:2804–2814. doi: 10.1128/mcb.18.5.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhl JD, Cook TA, Gebelein B. Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev Biol. 2010;343:154–166. doi: 10.1016/j.ydbio.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Xu L, Lee J, Xu T. Drosophila atrophin homolog functions as a transcriptional corepressor in multiple developmental processes. Cell. 2002;108:45–56. doi: 10.1016/s0092-8674(01)00630-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.