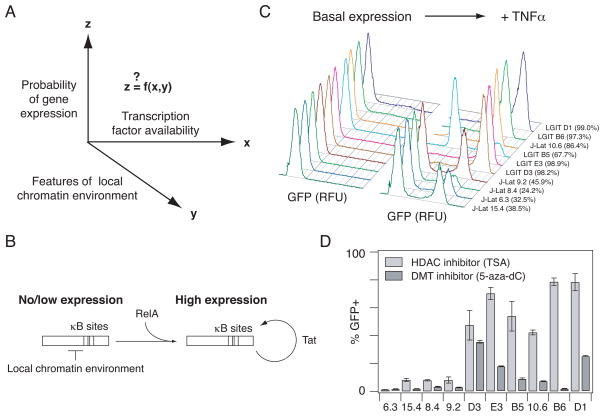
Fig. 1.
In vitro models of HIV gene expression provide an experimental system to study RelA-mediated gene expression in a range of chromatin environments. (A) There is general interest in how gene expression probability varies as a function of transcription factor availability and quantitative features of the local chromatin environment. (B) Schematic describing RelA-mediated gene expression in the HIV vectors before and after the Tat-mediated positive feedback loop is activated. (C) Representative flow cytometry histograms of GFP expression for the panel of clones each infected with a single integration of an inactive HIV provirus under basal conditions (left) and after stimulation with TNFα (20 ng/ml) for 48 hours (right). Percentage of TNFα-activated cells is indicated in parentheses. Clones are ordered according to increasing basal gene expression. (D) Infected clonal populations were stimulated with 400 nM TSA for 24 hours (light gray bars) or 5 μM 5-aza-dC for 48 hours (dark gray bars). Experiments were performed in biological triplicate. Data are presented as the mean ± standard deviation.