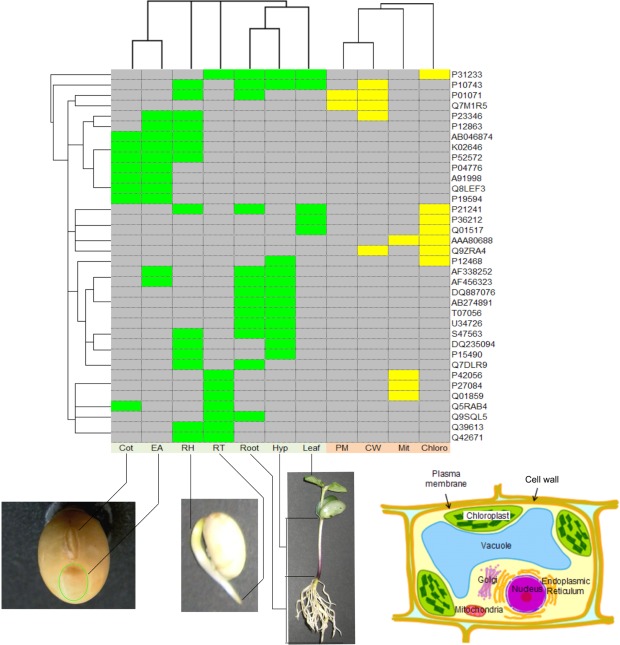
FIGURE 2.
Clustering results for 36 proteins identified in more than one organ and/or subcellular compartment. Accession numbers for the proteins are indicated on the right-hand side. Colored boxes indicate an identified protein found in the organ (green) or a subcellular compartment (yellow). Samples from seven organs, Cot (cotyledon), EA (embryonic axis), RH (radicle plus hypocotyl), RT (root tip), Root, Hyp (hypocotyls), and Leaf, and four subcellular compartments, PM (plasma membrane), CW (cell wall), Mit (mitochondrion), and Chloro (chloroplast), were investigated. The samples of Cot, EA, RH, RT, Root, Hyp, and Leaf were extracted 0, 0, 2, 3, 7, 7, and 7 days after seedling emergence, respectively. The samples comprising PM, CW, Mit, and Chloro were extracted 3, 4, 4, and 7 days after seedling emergence, respectively. Clustering was conducted based on the identification of corresponding proteins, as identified (1) or not-identified (0), separately for organs and subcellular compartments. Hierarchical clustering was performed using Gene Cluster 3.0 (de Hoon et al., 2004) with Euclidean distance and centroid linkage method. The resulting clusters were visualized using JAVA TREEVIEW (Saldanha, 2004).