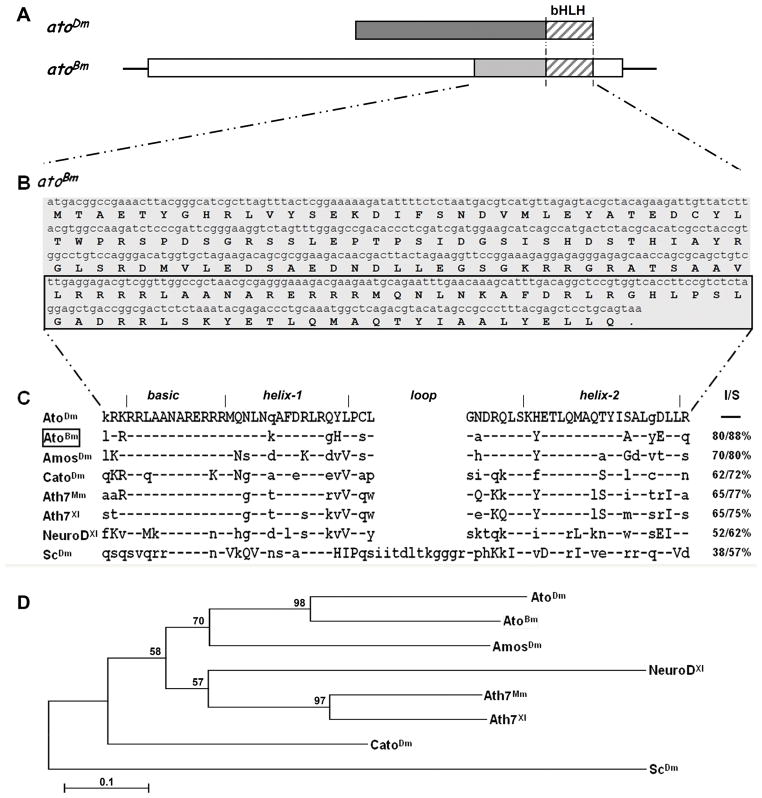
Figure 1. Identification of the atonal gene in Bombyx mori.
A) Schematic drawing of the transcription unit for atoBm . Open boxes represent 5′ and 3′ UTR, shaded boxes represent ORF with the stripes marking the region encoding the bHLH. Structure of the atoDm transcript is shown above for comparison. The two proteins are 80% identical and 88% conserved in the bHLH region (striped boxes), but not conserved elsewhere. See Supplementary Figure 1 for more details. B) Full length ORF and translation of atoBm. The bHLH domain is marked by the box. C) Sequence alignment of the bHLH domains of selected proneural proteins (Mm=Mus musculus; Xl=Xenopus laevis). Conservation is given as I=Identities (%identical aa) and S=Similarity (% identical + similar aa) as compared to AtoDm. Residues identical to AtoDm are marked by a hyphen (−), conserved changes are in upper case, nonconserved changes are in lower case. GenBank accession numbers are as follows: AtoDm = NP731223; AtoBm = HQ888870; AmosDm = NP477446; CatoDm = NP477344; Ato7Mm = NP058560; Ato7Xl = NP001079289; NeuroDXl = 1096595; ScDm = NP476803. D) Phylogenetic tree showing the relationship of atonal homologs from different species. The bootstrap 50% majority-rule consensus trees were made with the maximum parsimony method (PAUP, v.4.0b10) using multiple alignments of amino acid sequences. Statistical support (percentage) for each node was evaluated by bootstrap analysis with 1,000 replicates.