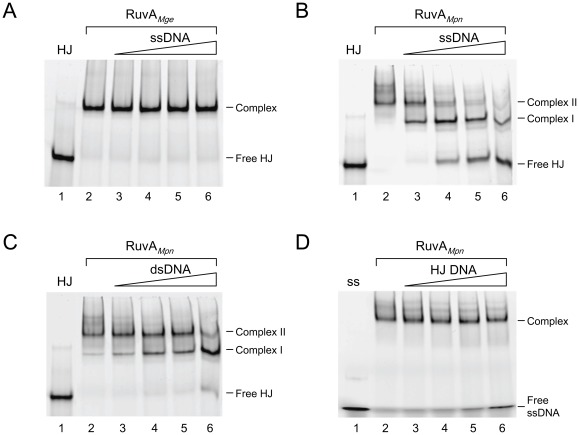
Figure 3. DNA binding preferences of RuvAMge and RuvAMpn.
(A) Binding of RuvAMge (3 µM) to HJ substrate HJ 1.1 (6-FAM-labeled on strand HJ11) in the presence of various concentrations of unlabeled ssDNA (oligonucleotide HJ11). The molar excess of unlabeled DNA over labeled DNA in the reactions was 0× (lane 2), 2.5× (lane 3), 5× (lane 4), 10× (lane 5) and 20× (lane 6). The protein was added as final component in the reactions. Protein was omitted from the reaction shown in lane 1. The positions of the free HJ substrate (Free HJ) and RuvAMge-HJ complexes (Complex) are indicated at the right-hand side of the gel. (B) Binding of RuvAMpn (3 µM) to HJ substrate HJ 1.1 (6-FAM-labeled on strand HJ11) in the presence of various concentrations of unlabeled ssDNA (oligonucleotide HJ11). The experiment was performed similarly as in (A). The two major RuvAMpn-HJ complexes (Complex I and II) are indicated at the right-hand side of the gel. (C) Binding of RuvAMpn (3 µM) to HJ substrate HJ 1.1 (6-FAM-labeled on strand HJ11) in the presence of various concentrations of unlabeled dsDNA (oligonucleotide HJ11/HJ11rv). The experiment was performed similarly as in (A). The two major RuvAMpn-HJ complexes (Complex I and II) are indicated at the right-hand side of the gel. (D) Binding of RuvAMpn (3 µM) to ssDNA (6-FAM-labeled oligonucleotide HJ11) in the presence of various concentrations of HJ DNA (HJ 1.1). The experiment was performed similarly as in (A). Protein was omitted from the reaction shown in lane 1. The positions of the unbound ssDNA (Free ssDNA) and RuvAMpn-ssDNA complex (Complex) are indicated at the right-hand side of the gel.