Table 2.
Scales of geographical population structure
A. Proportion of variation distributed among populations, 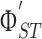 , at different spatial scalesa , at different spatial scalesa
| |||||
|---|---|---|---|---|---|
| Species | Marker | Between islands | Within Maui Nui | Within Big Island | Within kīpuka region |
| L1 Th. grallator | Allozymes | 0.46*** | 0.37*** | 0.19*** | – |
| CO1/16S/ND1 | 0.69*** | 0.56*** | 0.30*** | 0.05*** | |
| L2 Ariamnes species | Allozymes | – | – | – | – |
| COI | 0.37*** | 0.57*** | 0.05 | – | |
| L3 T. anuenue & T. kikokiko | Allozymes | 0.36*** | – | – | – |
| COI | 0.98*** | – | 0.23*** | 0.041*** | |
| L4 T. brevignatha & T. macracantha | Allozymes | – | 0.46*** | – | – |
| COI | 0.93*** | 0.76*** | 0.16* | 0.000 | |
| L5 T. quasimodo | Allozymes | 0.06*** | 0.15*** | 0.34*** | – |
| COI | 0.74*** | – | 0.09*** | 0.037*** | |
B. Comparison of genetic variance among populations, 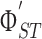 , at different scales for COI data. Includes among 4 lineages of habitat specialists (L1–L4, excluding L5 T. quasimodo and L6 T. hawaiensis) , at different scales for COI data. Includes among 4 lineages of habitat specialists (L1–L4, excluding L5 T. quasimodo and L6 T. hawaiensis) | ||
|---|---|---|
| Species set 1 (mean) | Species set 2 (mean) | Resultsb |
| 1. Within Maui (0.63) | >Within Big Island (0.18) | t = 4.13, p < .027* |
| 2. Within Big Island (0.18) | >Among kīpuka (0.03) | t = 10.6, p < .004** |
| 3. Between islands (0.74) | >Within Big Island (0.18) | t = 3.60, p < .035* |
| 4. Between islands (0.74) | >Within Maui (0.63) | t = 0.44, p < .35 |
| 5. Between islands mtDNA (0.74) | >Between islands nuclear (0.41) | Insufficient sample size |
| C. N e m, estimated relative gene flow among populations | ||||
|---|---|---|---|---|
| Species | Marker | Within Maui Nui | Within Big Island | Within kīpuka region |
| L1 Th. grallator | Allozymes | 2.6–8.1 | 3.3–7.1 | – |
| CO1/16S/ND1 | 0.6–2.1 | 1.9–4.2 | – | |
| L2 Ariamnes species | Allozymes | – | – | – |
| COI | – | 0.3–5.3 | – | |
| L3 T. anuenue & T. kikokiko | Allozymes | – | – | – |
| COI | – | 11.7 | 5.0 | |
| L4 T. brevignatha & T. macracantha | Allozymes | 5.6 | – | – |
| COI | 0.3 | 0.3–7.0 | – | |
| L5 T. quasimodo | Allozymes | 6.4 | 6.3–6.5 | – |
| COI | – | 0.3–13 | 4.9 | |
| L6 T. hawaiensis | Allozymes | – | – | – |
| COI | – | 12.0 | – | |
aSee text for description of analyses and calculations, * p < 0.05, *** p < 0.001
bArc-sin square-root transformed before analysis, df = 2, one-tailed t test, * p < 0.05, ** p < 0.01
