Figure 3.
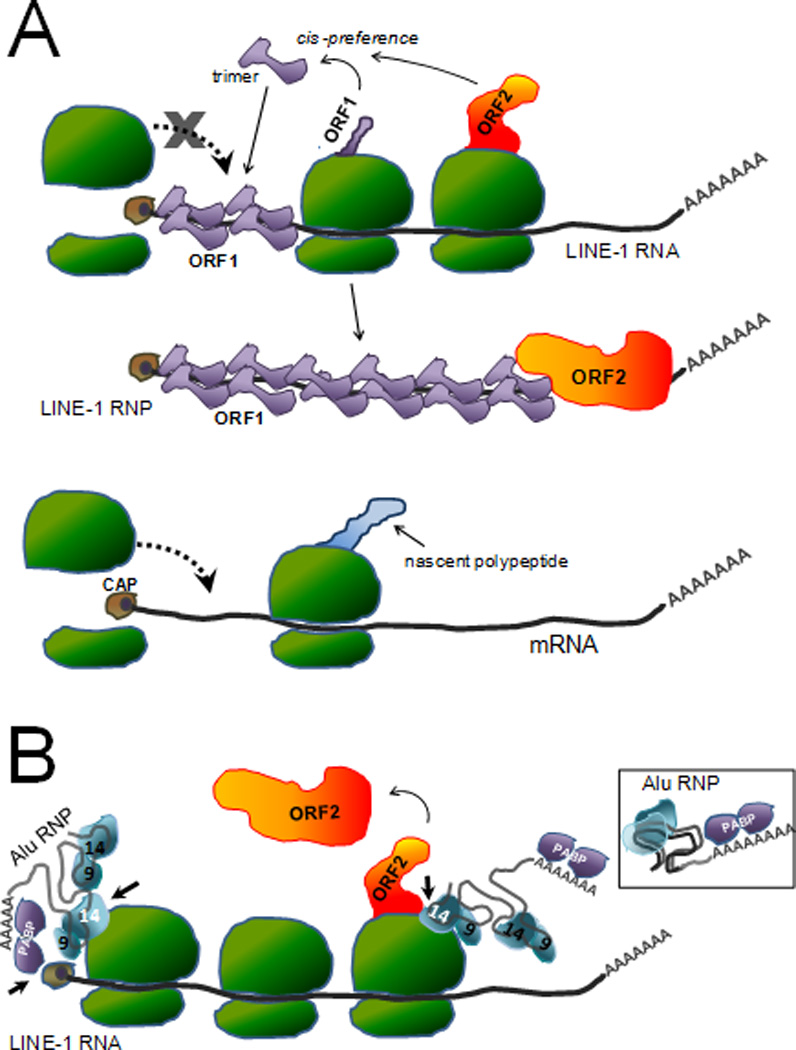
Proximity as a requirement for efficient retrotransposition. A. Proposed model explaining the reduced efficiency of retrotransposition of non-L1 mRNAs (retropseudogenes generation). Schematic representation of a full-length capped L1 transcript in a polyribosomal complex undergoing translation. As the L1 ORF1 protein is translated, the ORF1 monomers interact forming a trimer. Due to the cis-preference shown by L1 it is expected that the ORF1 trimer will bind the L1 RNA that generated it, likely driven by proximity. Although ORF2p is made in lower amounts, it also is thought to preferentially interact with the same L1 transcript. The increased number of ORF1p likely coats the L1 RNA, possibly preventing the reassembly of the ribosome and contributing to the “escape” from the translational machinery, allowing for the newly formed L1 RNP to progress through the retrotransposition cycle. In contrast, because mRNA from cellular genes will be undergoing translation in their own polyribosomes, these transcripts will be physically restricted from coming in contact with the L1 proteins. The lack of the ORF1p interaction with the mRNA will enable ribosomes to continue to assemble and continue translation, until the mRNA is targeted to the degradation pathway. [The CAP complex is represented as a brown circle at the 5’ end of the transcript. Ribosomes are shown in green, and the L1 ORF1 and ORF2 proteins are shown in purple and orange respectively.] B. Proposed model for Alu sequestration of ORF2 protein. Due to the lack of coding sequences, Alu transcripts are not engaged by the translation machinery. Thus the Alu transcript can exist as a “free” ribonucleoprotein (RNP) complex (see inset). The inset shows a liberal representation of the potential folding of the two 7SL derived monomers to form the propeller structure and the interaction with the SRP9 and SRP14 proteins (depicted in blue): only SRP9p is shown. In addition to SRP9/14p, the Poly-A binding protein (PABP, depicted in purple) is thought to interact with the A-tail of the Alu RNA. PABP is shown interacting with the 3’ A-tail of the Alu RNA. The structure is likely to fold to form a compact RNP. The bound SRP14p and PABP may play a role in targeting Alu RNPs to the ribosomes. SRP14p interacts with ribosome through direct contacts with amino acids located in its carboxy-terminus. The PABP bound to the Alu A-tail may also help direct the Alu RNP to the translating L1 transcript by interacting with the cap complex (brown circle) of the L1 RNA. These interactions (indicated by short black arrows) may favor the localization of Alu RNPs to translating ribosomes to increase their probability of coming in close proximity to newly synthesized ORF2p (in orange). For illustrative purposes, the L1 transcript bound to ribosome is shown as a linear molecule. However, the cap complex of the L1 RNA likely interacts with the PABP present in its own A-tail circularizing the RNA.
