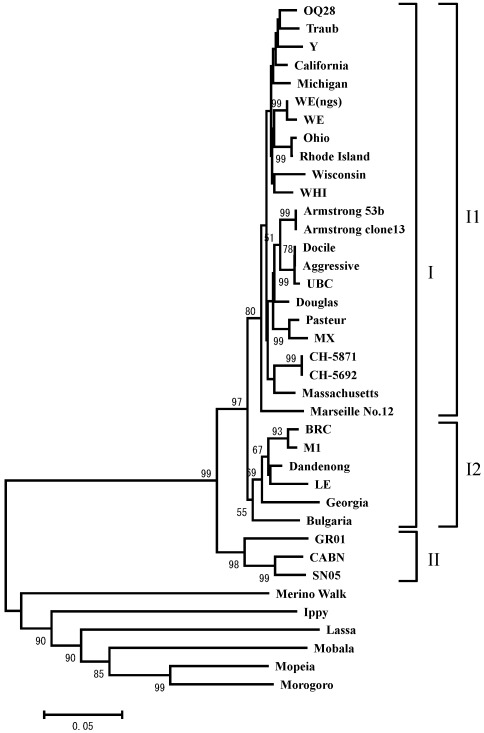
Figure 1.
Phylogenetic comparison of LCMV strains OQ28, BRC, and WE(ngs) to other Old World arenaviruses. The phylogenetic tree was based on amino acid sequences of NP coding region available in the GenBank database and was constructed with the neighbor-joining method by using MEGA version 4. The phylogenetic tree is based on 559-residue amino acid sequences of the NP coding region, although strain Y lacks the first 64 residues and strain LE lacks the first 195 amino acids. The numbers at the nodes are bootstrap values based on 1000 replications. A value greater than 50 is considered significant for a branch point. The scale bar represents 0.05 substitutions per amino acid position.