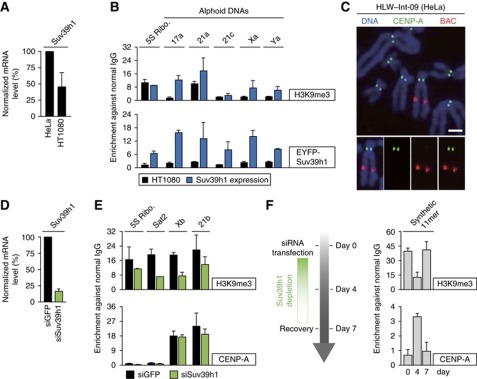
Figure 2.
Suv39h1, histone H3K9 tri-methylase, negatively regulates ectopic CENP-A assembly. (A) Suv39h1 expression level. Total RNA was purified from each cell line, reversely transcribed and quantified by real-time PCR. Suv39h1 mRNA amounts were normalized by HPRT transcripts. Both Suv39h1 and HPRT genes are on X chromosome. (B) Exogenous Suv39h1 expression induced H3K9me3 modification on centromeric alphoid DNAs in HT1080 cells. EYFP-tagged Suv39h1 gene was transfected and cells were harvested more than four weeks after transfection. ChIP was carried out with normal IgG and a set of indicated antibodies. Primer sets shown at the top were used for quantitative PCR. Error bars, s.d. (n=2). (C) Examples of no ectopic CENP-A assembly in HeLa integration cell (HLW-Int-09). Mitotic cells were spread on cover glass, and stained with DAPI (blue), anti-CENP-A antibody (green), and BAC DNA probe (red). Scale bar, 5 μm.(D) Depletion of Suv39h1 with siRNA. siRNAs for the GFP gene (siGFP; control) or Suv39h1 (siSuv39h1) was transfected to HLW-Int-09 cell. Total RNA was purified and quantified by real-time PCR. Suv39h1 mRNA levels were normalized by HPRT RNA. Vertical axis indicates relative Suv39h1 mRNA level against a negative control (siGFP). Error bar, s.d. (n=3). (E, F) ChIP assay was carried out with HLW-Int-09 cells treated by siGFP or siSuv39h1. Normal IgG and a set of different antibodies were used for ChIP. Indicated primer sets were used for quantitative PCR (top). Error bars in panel E, s.d. (n=2). (F) HLW-Int-09 cells were harvested at three time points, 0, 4 and 7 days after siSuv39h1 transfection and used for ChIP analysis. Error bars, s.d. (n=3).