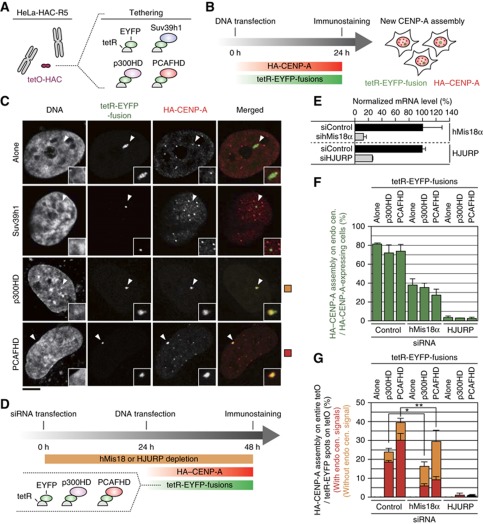
Figure 4.
HAT tethering on tetO-HAC induced expansion of newly synthesized CENP-A assembly through HJURP. (A) A HAC cell line (HeLa-HAC-R5) was transfected with a set of tetR-EYFP-fusion expressing vectors. (B) Timetable for the experiment. HA-tagged CENP-A expression vector (pCDNA5-HA-CENP-A) was co-transfected with tetR-EYFP-fusion expressing vector. (C) Representative images of newly synthesized CENP-A assembly. Cells were stained with DAPI, anti-GFP (recognize EYFP; green) and anti-HA (red). Arrowheads indicate tetO-HAC position. Scale bar, 5 μm. (D) Schematic timetable for gene depletion and new CENP-A assembly assay. HeLa-HAC-R5 cells were firstly transfected with siRNA. After 24 h incubation, HA-CENP-A and a set of tetR-EYFP-fusion expression vectors were co-transfected. Cells were stained with DAPI, anti-GFP and anti-HA. (E) hMis18α or HJURP depletion using siRNA. siRNAs for hMis18α (sihMis18α) and for HJURP (siHJURP) as well as for a negative control (siControl: siNegative, ambion) were used for transfection. Total RNA was purified two days after transfection and quantified by real-time PCR. hMis18α or HJURP mRNA levels were normalized by HPRT transcripts. Horizontal axis indicates relative hMis18α or HJURP mRNA level against a negative control (siControl). Error bar, s.d. (n=3). (F) HA-CENP-A assembly frequency on endogenous centromere was counted in each sample (n?100). Error bar, s.d. (n=3). (G) A frequency of expanded HA-CENP-A assembly induced by HAT tethering (example is shown in panel C bottom) was counted in each sample (n?100). Error bar, s.d. (n=3). Column colors indicate subpopulations of cells, which had CENP-A assembly at endogenous centromere (red) and had no assembly (orange). *P-values of t-test are 0.001 (red column) and 0.017 (orange column). **P-values of t-test are 0.006 (red column) and 0.033 (orange column).