Table 1.
Identification and quantitative analysis of early salt-responsive proteins in wild type and OSRK1 transgenic rice roots
| Spota | Protein Identification | Matched peptide |
Sequence Coverage (%) |
Mrb (Ex/Tr) |
NCBI Accession Number |
Fold changec | Protein ratiod (OSRK1/WT) |
Express Graphe |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | OSRK1 | ||||||||||
| 3 h | 7 h | 3 h | 7 h | ||||||||
| Translation and transcription | |||||||||||
| 5i | Elongation factor Tu | 9 | 26.3 | 43.7/48.4 | gi|21685576 | ndf | 7.32 | 2.47 | 1.19 | Ig | 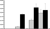 |
| 12i | Putative translation elongation factor eEF-1 beta chain |
5 | 35.3 | 30.0/23.8 | gi|113612079 | 0.97 | 2.00 | 1.92 | 0.55 | 1.25 | 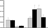 |
| 323 | Glycine-rich RNA-binding protein | 6 | 74.0 | 13.1/15.5 | gi|115441831 | 1.02 | 0.38 | 1.14 | 0.85 | 0.93 | 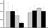 |
| 383 | Translation initiation factor 5A | 2 | 25.0 | 17.7/17.4 | gi|113611710 | nd | 2.84 | 1.04 | 0.62 | Ig | 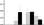 |
| Amino acid and purine metabolism | |||||||||||
| 26 | Putative inosine monophosphate dehydrogenase | 2 | 4.70 | 57.4/51.9 | gi|215694434 | 0.38 | 1.25 | 0.89 | 0.88 | 1.05 | 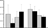 |
| 27 | Aspartate-semialdehyde dehydrogenase | 3 | 10.9 | 40.1/40.1 | gi|113549818 | 2.11 | 3.05 | 1.15 | 0.80 | 5.46 | 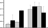 |
| 67 | Aspartate aminotransferase | 4 | 13.0 | 40.6/44.8 | gi|29468084 | 1.48 | 4.51 | 0.87 | 0.97 | 3.64 | 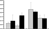 |
| 68 | Putative isovaleryl-CoA dehydrogenase | 1 | 2.70 | 41.5/44.5 | gi|113578072 | 0.88 | 1.89 | 0.91 | 0.89 | 2.53 | 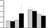 |
| 200 | Glutamine synthetase | 4 | 10.4 | 41.4/39.2 | gi|124052115 | 1.51 | 1.37 | 0.98 | 1.02 | 1.72 | 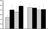 |
| 259 | Methylmalonate semi-aldehyde dehydrogenase | 5 | 10.5 | 18.1/57.2 | gi|113610618 | 0.46 | 0.60 | 1.22 | 0.67 | 0.27 | 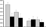 |
| 314 | Aspartate aminotransferase | 6 | 18.8 | 42.2/44.9 | gi|215768565 | 1.13 | 3.14 | 0.87 | 1.06 | 3.08 | 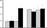 |
| 314 | Glutamate dehydrogenase | 7 | 21.4 | 42.2/44.3 | gi|33242905 | 1.13 | 3.14 | 0.87 | 1.06 | 3.08 | |
| Detoxyfying enzymes | |||||||||||
| 51 | Glutathione reductase, cytosolic | 11 | 24.8 | 55.6/53.4 | gi|113538016 | 0.63 | 2.80 | 0.89 | 1.01 | 1.82 | 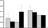 |
| 112i | Gutathione S-transferase | 4 | 25.6/25.6 | gi|31433227 | 1.67 | 0.72 | 1.35 | 1.51 | 0.28 | 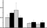 |
|
| 116 | Ascorbate peroxidase | 5 | 33.6 | 25.8/27.1 | gi|50920595 | 1.79 | 1.13 | 0.80 | 1.10 | 2.40 | 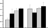 |
| 125 | Glutathione S-transferase II | 6 | 32.1 | 26.0/24.0 | gi|3746581 | 2.05 | 1.42 | 0.94 | 0.90 | 2.63 | 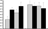 |
| 160i | Ascorbate peroxidase | 10 | 46.0 | 24.0/27.1 | gi|1321661 | 1.35 | 0.38 | 1.51 | 0.71 | 0.41 | 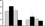 |
| 198 | Quercetin 3-O-methyltransferase | 2 | 6.8 | 41.4/39.7 | gi|113623000 | 1.57 | 1.19 | 1.06 | 0.84 | 1.83 | 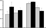 |
| 200 | Peroxidase | 3 | 8.6 | 41.4/37.8 | gi|257657027 | 1.51 | 1.37 | 0.98 | 1.02 | 1.72 | 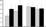 |
| 202 | Similar to glyoxalase II | 6 | 28.2 | 29.9/33.1 | gi|113533338 | 1.07 | 1.98 | 0.94 | 0.90 | 1.47 | 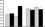 |
| 206 | Glyoxalase I | 10 | 51.5 | 33.9/32.5 | gi|113623141 | 3.57 | 1.73 | 0.84 | 1.00 | 4.46 | 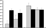 |
| 213 | Thioredoxin Type H | 7 | 50.8 | 13.6/13.9 | gi|82407383 | 1.94 | 0.87 | 1.01 | 0.98 | 3.20 | 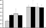 |
| 265i | Similar to glyoxalase II | 7 | 32.5 | 32.5/33.2 | gi|113533338 | 0.59 | 1.52 | 0.68 | 0.74 | 1.90 | 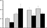 |
| 310 | Ascorbate peroxidase | 5 | 38.2 | 25.6/27.1 | gi|257707656 | 0.79 | 0.34 | 1.85 | 4.40 | 0.10 | 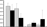 |
| 351 | Ascorbate peroxidase | 4 | 24.8 | 26.7/27.1 | gi|1321661 | 1.98 | 0.65 | 1.00 | 0.97 | 1.35 | 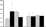 |
| 390 | Aldehyde dehydrogenase | 3 | 8.0 | 54.6/59.2 | gi|8163730 | 3.16 | 3.10 | 0.85 | 1.25 | 10.85 | 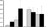 |
| Glycolysis and other carbohydrate metabolism related proteins | |||||||||||
| 61 | Fructose -bisphosphate aldolase | 3 | 9.8 | 29.3/38.8 | gi|790970 | 0.26 | 0.94 | 0.69 | 0.81 | 0.82 | 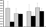 |
| 67 | Phosphoglycerate kinase | 3 | 11.0 | 40.6/42.3 | gi|113596357 | 1.48 | 4.51 | 0.87 | 0.97 | 3.64 | 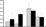 |
| 76i | Dihydrolipoamide dehydrogenase precursor | 5 | 13.5 | 60.6/52.6 | gi|113532449 | nd | 16.87 | 0.87 | 1.54 | Ig | 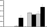 |
| 77 | Transketolase | 7 | 10.6 | 76.8/80.0 | gi|227468492 | 2.14 | 15.09 | 1.02 | 0.98 | 18.68 | 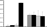 |
| 79 | Similar to enolase | 6 | 15.9 | 52.0/50.7 | gi|115478881 | 1.90 | 0.78 | 0.80 | 0.74 | 3.14 | 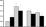 |
| 116 | Triose phosphate isomerase | 13 | 79.1 | 25.8/27.6 | gi|553107 | 1.79 | 1.13 | 0.80 | 1.10 | 2.40 | 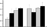 |
| 275 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase | 8 | 27.2 | 70.8/60.8 | gi|257353836 | 1.45 | 2.56 | 0.80 | 0.84 | 2.07 | 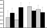 |
| 277 | 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase | 3 | 12.2 | 70.8/60.8 | gi|257353836 | 1.26 | 2.83 | 0.86 | 0.88 | 2.33 | 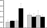 |
| 279i | Enolase | 4 | 14.4 | 57.0/47.9 | gi|113548027 | 1. 03 | 1.70 | 0.72 | 0.80 | 1.41 | 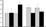 |
| 292 | Phosphoglyceromutase | 9 | 20.8 | 69.9/60.7 | gi|257353838 | 1.03 | 2.13 | 0.93 | 1.04 | 2.66 | 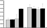 |
| 293 | Phosphoglyceromutase | 12 | 28.1 | 69.9/60.7 | gi|257353838 | 1.50 | 1.79 | 0.87 | 1.02 | 1.90 | 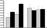 |
| 315 | Glyceralde-3-phosphate dehydrogenase | 4 | 16.3 | 39.1/36.5 | gi|968996 | 0.99 | 2.14 | 0.91 | 1.10 | 2.15 | 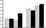 |
| 318 | Glyceralde-3-phosphate dehydrogenase | 4 | 16.3 | 38.7/36.4 | gi|968996 | 1.17 | 2.68 | 1.03 | 1.01 | 3.06 | 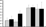 |
| 353 | Triose phosphate isomerise | 4 | 20.9 | 22.0/27.0 | gi|169821 | 0.83 | 0.62 | 1.16 | 0.67 | 0.32 | 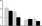 |
| 385 | Phosphoglycerate kinase | 5 | 16.7 | 45.4/42.2 | gi|113596357 | nd | 4.57 | 1.03 | 1.10 | Ig | 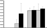 |
| Proteolytic enzymes | |||||||||||
| 112i | Proteasome subunit beta type 2 | 5 | 26.4 | 25.6/23.4 | gi|17380213 | 1.67 | 0.72 | 1.35 | 1.51 | 0.28 | 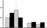 |
| 134 | Oryzacystatin | 3 | 23.5 | 13.3/11.4 | gi|1280613 | 2.10 | 0.46 | 0.87 | 1.00 | 0.22 | 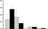 |
| 204 | Proteasome subunit alpha type 2 | 7 | 34.6 | 26.2/27.6 | gi|259443357 | 0.59 | 1.88 | 0.89 | 1.15 | 1.22 | 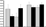 |
| 351 | Proteasome subunit alpha type 2 | 4 | 16.6 | 26.7/25.8 | gi|259443327 | 1.98 | 0.65 | 1.00 | 0.97 | 1.35 | 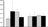 |
| Heat shock proteins | |||||||||||
| 112i | Putative chaperonin21 precursor | 5 | 26.2 | 25.6/26.3 | gi|51090748 | 1.67 | 0.72 | 1.35 | 1.51 | 0.28 | 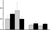 |
| 274 | Chaperonin CPN60-1, mitochondrial precursor | 10 | 17.5 | 64.1/61.0 | gi|113547409 | 1.17 | 2.77 | 0.88 | 1.12 | 1.49 | 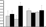 |
| 276i | DnaK-type molecular chaperone precursor | 2 | 4.6 | 75.0/70.4 | gi|257307253 | 1.26 | 3.35 | 0.76 | 1.00 | 2.78 | 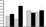 |
| 278i | DnaK-type molecular chaperone precursor | 6 | 14.5 | 72.9/70.4 | gi|257307253 | 2.35 | 2.38 | 0.66 | 1.47 | 4.47 | 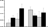 |
| Lipid biosynthesis | |||||||||||
| 258 | Putative enoyl-ACP reductase | 3 | 15.7 | 35.4/39.1 | gi|113623526 | 1.32 | 2.41 | 1.13 | 1.01 | 2.92 | 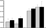 |
| 69 | Putative acetyl-CoA C-acetyltransferase | 6 | 19.2 | 43.1/44.1 | gi|113630918 | 1.28 | 3.46 | 0.80 | 0.99 | 6.40 | 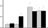 |
| 82 | Putative inorganic pyrophosphatase | 5 | 21.4 | 31.8/33.0 | gi|113537770 | 0.88 | 0.41 | 0.97 | 0.99 | 0.42 | 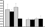 |
| 177 | Nucleoside diphosphate kinase from rice | 2 | 16.6 | 14.5/16.8 | gi|113639936 | 0.39 | 0.81 | 0.88 | 0.83 | 1.14 | 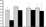 |
| 227i | Putative inorganic pyrophosphatase | 5 | 13.9 | 32.1/33.0 | gi|113537770 | 1.70 | 0.41 | 0.92 | 1.30 | 0.44 | 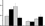 |
| Stress related proteins | |||||||||||
| 124 | Formate dehydrogenase, mitochondrial precursor | 13 | 47.2 | 42.4/41.2 | gi|21263611 | 0.64 | 7.24 | 0.78 | 0.90 | 4.13 | 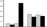 |
| 134 | 15 kda Organ-specific salt-induced protein | 2 | 23.4 | 13.3/15.2 | gi|256638 | 2.10 | 0.46 | 0.87 | 1.00 | 0.22 | 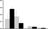 |
| 214 | 15 kda Organ-specific salt-induced protein | 7 | 77.2 | 13.4/15.2 | gi|256638 | 4.05 | 0.33 | 0.87 | 1.12 | 0.48 | 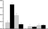 |
| 227i | 15 kda Organ-specific salt-induced protein | 6 | 64.8 | 32.1/15.2 | gi|256638 | 1.70 | 0.41 | 0.92 | 1.30 | 0.44 | 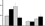 |
| 386 | Pathogen-related protein | 4 | 21.3 | 16.5/17.2 | gi|16589076 | 4.29 | 2.28 | 1.30 | 0.38 | 6.70 | 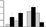 |
| Signal transduction related proteins | |||||||||||
| 225 | Calreticulin | 3 | 5.9 | 33.4/47.9 | gi|6682833 | 0.75 | 0.43 | 1.47 | 1.26 | 0.26 | 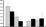 |
| Unkwown proteins | |||||||||||
| 392 | Hypothetical protein | 3 | 10.7 | 25.8/27.4 | gi|14192878 | 5.65 | 4.00 | 0.79 | 0.96 | 18.95 | 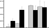 |
aSpot numbers correspond to the spots in Figure 2 and Figure 3
bMolecular weight (kDa) of protein spots estimated from gel analysis (Ex) and theoretical molecular weight of identified protein (Tr)
cFold change of each spot after NaCl treatment for 3 h or 7 h, calculated from the mean spot volumes in control and NaCl treated gel groups
dSpot-volume ratio of WT to OSRK1, calculated from the mean spot volumes in 3 h control of WT and OSRK1 groups
eRelative expression graphs of protein spots after NaCl treatment in WT and OSRK1. Spot volumes are analyzed by Progenesis software. The left four bars indicate spot volume of WT root and right four bars indicate spot volumes of OSRK1. From left to right, each bar indicate spot volume of 3 h control, 3 h NaCl treated, 7 h control and 7 h NaCl treated gel groups. Values are means ± SE. Error bars from three spots in three independent gels
fnd: not detected
g I: spots were not detected in WT groups, but appeared in OSRK1 groups
hProteins not identified from the database
i Salt-responsive proteome identified from OSRK1 transgenic rice roots
