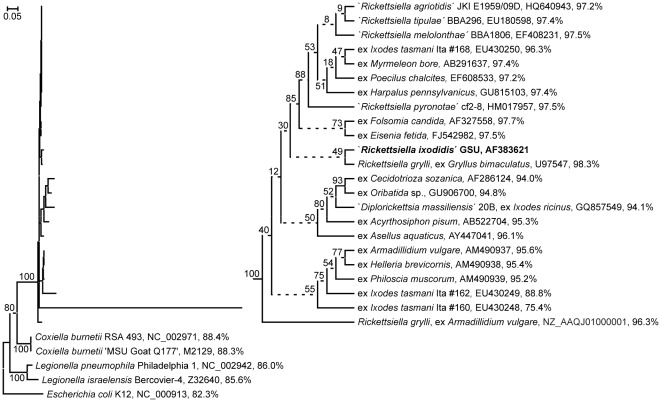
Figure 1. Bacterial ML phylogeny generated from 16 S ribosomal RNA encoding sequences.
Terminal branches are labelled by genus and species, pathotype, strain and/or original host designations, GenBank accession numbers as well as pairwise nucleotide sequence identity percentages as calculated from a p-distance matrix with respect to the ‘Rickettsiella ixodidis’ GSU sequence. Numbers on internal branches indicate bootstrap support values; branches that do not receive >90% bootstrap support are represented by dashed lines. The phylogram has been rooted using Escherichia coli as outgroup. The size bar corresponds to 5% sequence divergence. To enhance resolution, the upper clade of the phylogram that comprises Rickettsiella-like bacteria, has been extended into a cladogram. GenBank accession numbers AM490937-39 and EU430248-50 designate partial 16 S rRNA gene sequences comprising only 40–70% of the complete 16 S rRNA marker sequence. The tree has been reconstructed from ClustalX aligned sequences; an essentially identical ML tree has been generated from a T-Coffee based nucleotide sequence alignment (not shown).