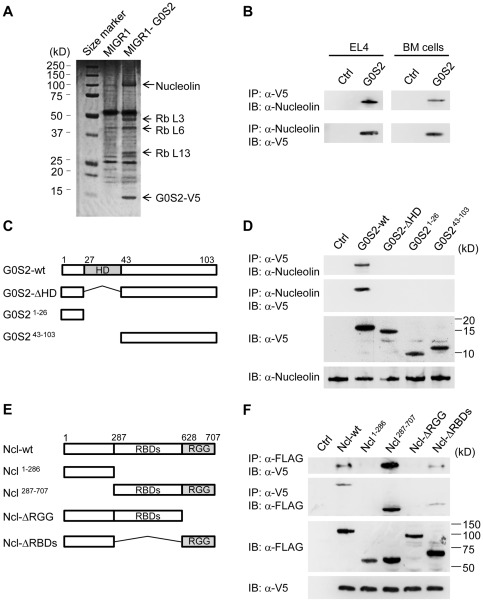
Figure 5. The hydrophobic domain of G0S2 interacts with the RGG domain of nucleolin.
Co-immunoprecipitation and proteomic analyses were used to identify G0S2 protein partners. (A) G0S2-interacting proteins were pulled down with anti-V5 antibody in EL4 cells transduced with the empty or G0S2-V5 tagged retrovirus. Bands from a SDS-PAGE gel stained with Coomassie Blue were cut out for identification by mass spectrometry. (B) Reciprocal co-immunoprecipitation of V5-tagged G0S2 and nucleolin in EL4 and BM cells transduced with the MIGR1 (Ctrl) or MIGR1-G0S2 (G0S2) retrovirus. (C) Diagram depicting the domains in the wild-type G0S2 protein and the deletion mutants G0S2-ΔHD, G0S21–26 and G0S243–103. (D) The interaction between endogenous nucleolin and ectopic V5-tagged G0S2 was analyzed in NIH3T3 cells transduced with the empty retrovirus (Ctrl) or retroviruses bearing wild-type G0S2 (G0S2-wt) or G0S2 mutant constructs (G0S2-ΔHD, G0S21–26 and G0S243–103). (E) Diagram depicting the domains of the wild-type nucleolin (Ncl) protein and the deletion mutants Ncl1–286, Ncl287–707, Ncl-ΔRGG and Ncl-ΔRBDs (FLAG-tagged). RGG, arginine-glycine-glycine-rich domain; RBD, RNA-binding domain. (F) Interactions between V5-tagged G0S2 and FLAG-tagged nucleolin constructs (Ncl-wt, Ncl1–286, Ncl287–707, Ncl-ΔRGG and Ncl-ΔRBDs). The data represent two independent experiments.