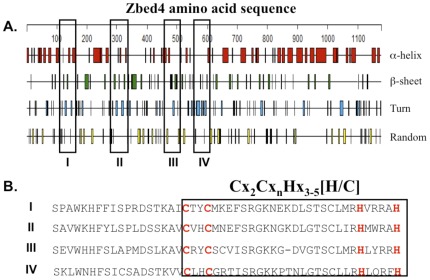
Figure 3. Prediction of secondary structures for Zbed4 using the Garnier-Robson algorithm.
A. Position of α-helices, β-sheets, turns and random amino acid sequences in the Zbed4 1,168 amino acid sequence. Predictions performed by two different programs, DNASTAR (Lasergene) and the FASTA Sequence Comparison web resource at the University of Virginia, gave the same results. Boxes I, II, III and IV indicate the position of BED zinc-finger motifs in Zbed4. B. The amino acid sequence of the four boxes in A show the specific signature of the BED-type zinc-fingers present in Zbed4 containing the characteristic cystines and histidines (red).