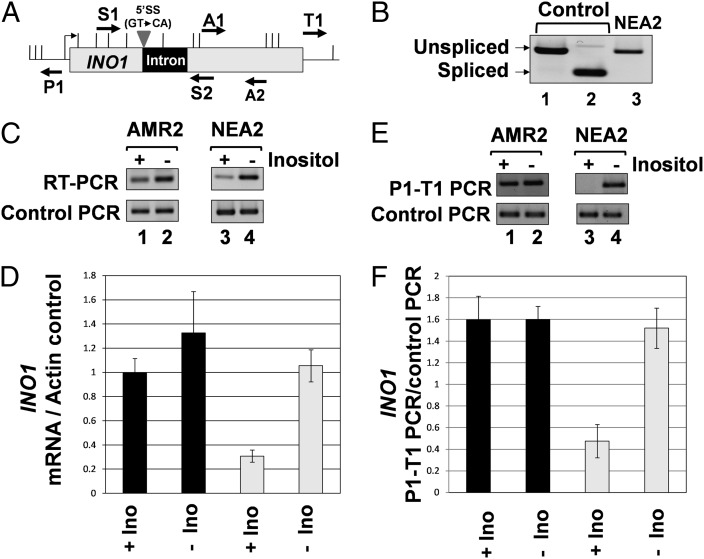
Fig. 2.
Splicing is required for intron-dependent enhancement of INO1 transcription and gene looping. (A) Schematic depiction of INO1 with a 5′ splice site mutated intron indicating the position of AluI and EcoRV restriction sites (vertical lines) and PCR primers used in RT-PCR and CCC analysis. The strain with the mutated 5′ splice site is NEA2, whereas the strain with the wild-type splice sites is AMR2 (B) ACT1 intron with a mutated 5′ splice site is not removed from INO1 pre-mRNA as indicated by the position of RT-PCR product obtained by using primers S1 and S2 flanking the intronic region of INO1 (lane 3). Control PCRs with and without intron were obtained by using genomic DNA from AMR2 and BY4733 as template using primers S1 and S2. (C) RT-PCR analysis of INO1 harboring a 5′ splice site mutated intron. The reverse transcription was performed by using oligo dT, and the cDNA obtained was PCR amplified by A1 and A2 primers. The control PCR represent the ACT1 mRNA levels that are used as a loading control, indicating that equal amount of template was present in each of the RT-PCRs. (D) Quantification of the data shown in C. (E) CCC analysis of INO1 containing a 5′ splice site mutated intron. The PCR products are indicated by the primers used for amplification. A P1-T1 PCR product indicates a looped configuration. The A1-A2 PCR represent the loading control, indicating that equal amounts of template DNA were present in each CCC PCR. (F) Quantification of the data shown in E.