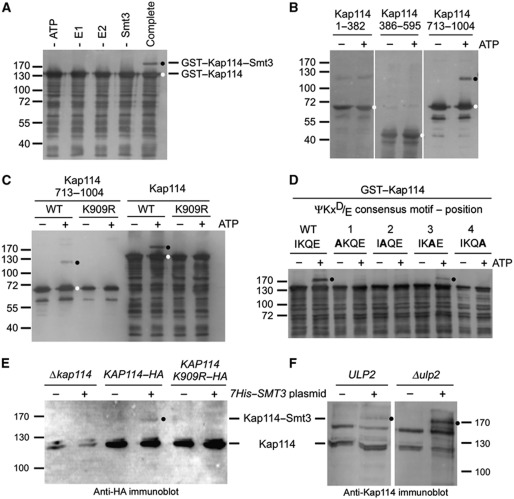
Figure 1.
Kap114 is sumoylated in vitro and in vivo on lysine residue 909. (A) Kap114 is sumoylated in vitro in the absence of an E3 ligase. Purified GST–Kap114 was incubated with 2 μg Aos1/Uba2 (E1), 0.75 μg Ubc9 (E2), 7.5 μg Smt3 (SUMO), and 5 mM ATP (complete mixture), as described in Materials and methods, at 30°C for 90 min. In control reactions, ATP, E1, E2, or Smt3 was omitted as indicated. (B) Kap114 is sumoylated at its C terminus. GST fusion proteins with Kap1141–382, Kap114386–595, and Kap114713–1004 were incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP at 30°C for 90 min. (C) Sumoylation of Kap114 occurs on lysine residue 909. GST fused to Kap114713–1004 or full-length Kap114 either unmutated (WT) or carrying the point mutation K909R was incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP. After sumoylation, GST fusion proteins were purified by pulldown assays using glutathione sepharose (A–C). (D) Sumoylation occurs at the ΨKxD/E consensus motif between residues 908 and 911 of Kap114. GST fusion proteins with full-length WT Kap114 or the point mutants I908A, K909A, Q910A, and E911A were incubated with E1, E2, and Smt3 in the presence or absence of 5 mM ATP at 30°C for 90 min. (E) Kap114 is sumoylated in vivo. Yeast cells deleted for KAP114 transformed with plasmids pRS426–KAP114–HA, pRS426–KAP114 K909R–HA, and YEp181–CUP–HIS7–SMT3 as indicated were grown in liquid cultures in the presence of copper sulphate for synthesis of His-tagged Smt3. (F) Sumoylation of Kap114 is increased in Δulp2 mutants. WT cells or ulp2 knockout cells transformed with plasmids pRS426–KAP114–HA and YEp181–CUP–HIS7–SMT3 were grown in liquid cultures in the presence of copper sulphate for synthesis of His-tagged Smt3. The cells were collected and resuspended in buffer containing 6 M guanidine–HCl. After glass bead lysis, cleared lysates were incubated with Ni-NTA agarose. Bound proteins were washed with buffer containing 8 M urea and eluted with SDS buffer (E, F). All proteins were separated by SDS–PAGE and detected by immunoblotting analysis using Kap114-specific affinity-purified antibodies (A–D, F) or anti-HA antibodies (E) and peroxidase-coupled secondary antibodies. Molecular weight markers are shown in kDa. Unmodified proteins are marked with a white dot, sumoylation products are labelled with a black dot.