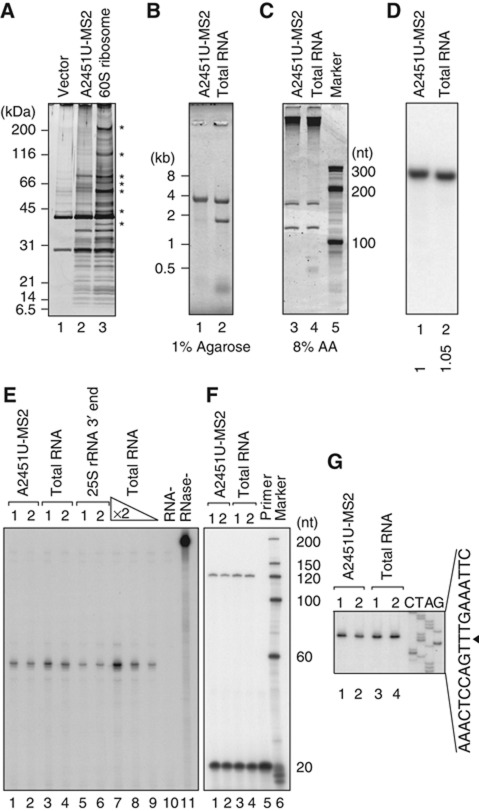
Figure 5.
Nonfunctional 60S particles that accumulated under proteasome-depleted conditions contained most ribosomal proteins and complete sets of all three rRNAs. (A–G) The protein and RNA compositions of nonfunctional ribosomes affinity purified with GST–MS2 from the RPT2 tet-off strain 4 h after transcriptional shut-off. GST pull-down was performed using ∼60S fractions. (A) The eluates were separated and silver stained. In lane 3, WT1–MS2-containing 60S ribosomal particles were purified from the wild-type strain (*non-ribosomal proteins). (B, C) Purified RNAs were separated and stained with SYBR Gold. In lanes 2 and 4, total RNA was purified from the wild-type strain. (D) Northern hybridization was performed using a proof which was designed to bind the 3′-most region of 25S rRNA (position +3251). The number below each lane shows the relative signal intensity. (E) An RNase protection assay was used to examine the 3′ ends of the 25S rRNAs. In lanes 5 and 6, the 3′ region of the 25S rRNA was transcribed in vitro and used. For lanes 7–9, total RNAs diluted two-fold were used. For lane 10, no target RNA was added to the reaction. Lane 11 is the no-RNase control. (F, G) Primer extension showed the 5′ ends of the 25S rRNAs. The primer used was designed to bind the region into which the tags were inserted (Supplementary Figure S7D). (G) The corresponding sequence for the sense strand of the 25S rDNA is shown. The 5′ end of the mature 25S rRNA is indicated by an arrowhead. Figure source data can be found with the Supplementary data.