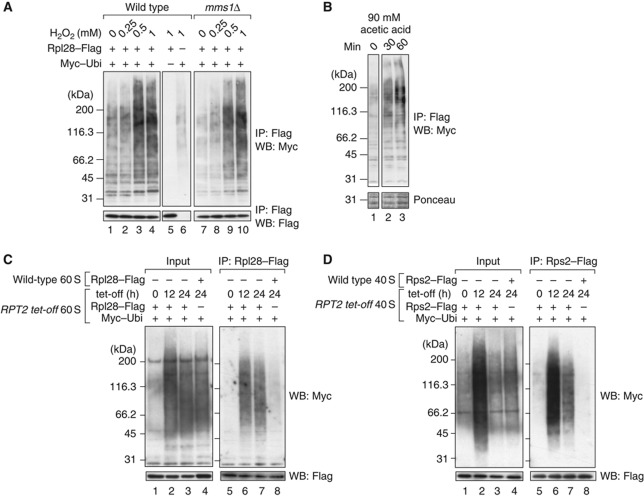
Figure 6.
Ribosomes are continuously ubiquitinated and degraded by proteasomes. (A, B) Ubiquitination signals induced by stress. (A) Wild-type and mms1Δ cells were grown in SD–glucose medium and treated for 2 h with 0, 0.25, 0.5, and 1 mM H2O2. In lane 5, an empty vector was used instead of pMyc–Ubi. In lane 6, another empty vector was used instead of pRpl28–Flag. (B) Wild-type cells were treated with acetic acid at a final concentration of 90 mM. Ribosomes were isolated at the indicated time points. (C) Ubiquitinated 60S ribosomal particles accumulated under Rpt2p-depleted conditions. The RPT2 tet-off strain was treated for the indicated times with 10 μg/ml Dox. The 60S ribosomal fractions were collected by 10–40% sucrose gradient sedimentation containing 40 mM EDTA. The 60S particles from the RPT2 tet-off strain were immunoprecipitated (lanes 5–7). Before the pull-down assay, untagged wild-type 60S fractions were mixed with the RPT2-tet-off strain-derived 60S fractions. To evaluate the non-specific binding of ubiquitinated proteins to ribosomes, as a control, 60S particles from the wild-type strain were immunopurified after they were mixed with the 60S fraction from RPT2 tet-off cells (lane 8). The ubiquitinated proteins in the mixture of 60S fractions are shown in lanes 1–4. (D) Ubiquitinated 40S ribosomal particles under Rpt2-depleted conditions. A similar set of experiments to (C) was performed using the 40S fractions and Rps2–Flag. Figure source data can be found with the Supplementary data.