Abstract
Genetic screens in simple model organisms have identified many of the key components of the conserved signal transduction pathways that are oncogenic when misregulated. Here, we identify H37N21.1 as a gene that regulates vulval induction in let-60(n1046gf), a strain with a gain-of-function mutation in the Caenorhabditis elegans Ras orthologue, and show that somatic deletion of Nrbp1, the mouse orthologue of this gene, results in an intestinal progenitor cell phenotype that leads to profound changes in the proliferation and differentiation of all intestinal cell lineages. We show that Nrbp1 interacts with key components of the ubiquitination machinery and that loss of Nrbp1 in the intestine results in the accumulation of Sall4, a key mediator of stem cell fate, and of Tsc22d2. We also reveal that somatic loss of Nrbp1 results in tumourigenesis, with haematological and intestinal tumours predominating, and that nuclear receptor binding protein 1 (NRBP1) is downregulated in a range of human tumours, where low expression correlates with a poor prognosis. Thus NRBP1 is a conserved regulator of cell fate, that plays an important role in tumour suppression.
Keywords: intestine, progenitor cell, Ras, tumour suppressor gene, WNT
Introduction
Perturbation of RAS, WNT or NOTCH signalling in the C. elegans vulva results in defects in vulval development, and many genetic screens have exploited this fact to identify important components of these pathways. In humans, the RAS, WNT and NOTCH pathways play an important role in orchestrating embryonic development and may be dysregulated in tumourigenesis. In the intestine, these pathways play critical roles in lineage specification and are essential for the normal formation of the bowel. Importantly, stem cells at the base of the intestinal crypt express the leucine-rich repeat-containing G-protein coupled receptor 5 (Lgr5) and are the progenitors from which all other cell types of the intestine (enterocytes, Paneth cells, goblet cells and enteroendocrine cells) are generated. Daughter cells of the Lgr5+ stem cells, called transit-amplifying cells, divide every 12–16 h generating some 300 cells per crypt per day and drive the replenishment of the intestinal epithelium, which is completely renewed every 5 days (Barker et al, 2009). Therefore, disruption of the fine balance between differentiation and proliferation by perturbing key signalling pathways has a profound effect on the ability of the intestine to regenerate and function.
Using RNAi in C. elegans, we performed a kinome-wide screen and identified the worm orthologue (H37N21.1) of the nuclear receptor binding protein 1 (NRBP1) as a novel enhancer of the RAS-induced multivulval phenotype (Muv) in a let-60(1046gf) gain-of-function mutant background. NRBP1 is located on human chromosome 2p23 and encodes a 535 amino-acid protein. It is ubiquitously expressed and highly conserved (Hooper et al, 2000). NRBP1 is a pseudokinase because it lacks critical catalytic residues in the kinase core, and carries a highly degraded ATP-binding site (Boudeau et al, 2006). In addition to its kinase-like domain, NRBP1 has a putative binding domain for Src homology-2-containing proteins, nuclear export signals, a nuclear localisation signal, a BC-box motif and a binding site for the myeloid leukaemia factor 1 (Gluderer et al, 2010). Here, we show that somatic deletion of Nrbp1 in the mouse causes a progenitor cell phenotype that disrupts the normal programme of cellular differentiation and proliferation along the intestinal crypt–villus axis. We also show that knockdown of NRBP1 cooperates with RasV12 to elicit transformation. Importantly, we show that loss of Nrbp1 increases the level of Sall4 and Tsc22d2 in the intestine where they associate with Nrbp1 and components of the ubiquitination machinery. In addition, we reveal that Nrbp1 is a tumour suppressor in mice and that NRBP1 is downregulated in a range of human cancers where low expression is associated with a poor clinical outcome. Thus, we identify critical roles for an uncharacterised gene through a combination of genetic screening in the worm and detailed functional analysis in the mouse and in human cells.
Results
Identification of H37N21.1 in a kinome-wide RNAi screen in let-60(n1046gf) worms
C. elegans that are homozygous for a gain-of-function allele of the worm Ras orthologue let-60(n1046gf) have a partially penetrant Muv phenotype with ∼60% of worms being Muv at 16 °C. The let-60(n1046gf) allele encodes an activating mutation in LET-60 that is functionally analogous to the codon 13 Gly>Glu mutation in RAS found in human tumours (Beitel et al, 1990). Multiple genetic screens have been carried out to identify genes whose loss suppresses the Muv phenotype in let-60(n1046gf) animals, and these have resulted in the isolation of many oncogenes acting in the RAS signalling pathway. Here, we use RNAi to identify genes whose loss enhances the Muv phenotype of let-60(n1046gf) reasoning that orthologues of these genes might function as tumour suppressors by perturbing the WNT, NOTCH or RAS pathways, which are critical to the formation of the worm vulva. This is the first such screen for enhancers of the Muv phenotype in let-60(n1046gf).
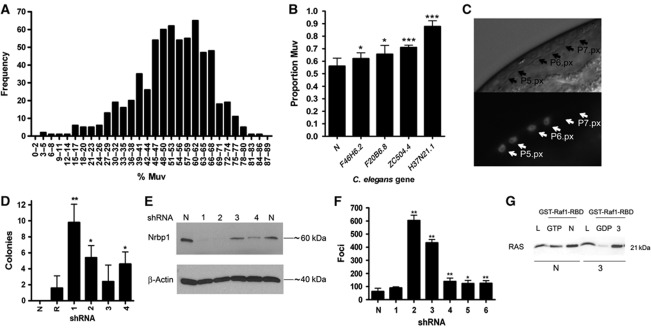
We adapted standard RNAi screening protocols (Fraser et al, 2000) to establish screening conditions in which we could robustly identify three previously characterised enhancers (egr-1, egl-27 and nhl-2) (Solari and Ahringer, 2000; Hammell et al, 2009) of the let-60(n1046gf) Muv phenotype. RNAi clones targeting all 656 genes in the worm genome with kinase homology were then screened (Figure 1A). For each gene, ∼500 worms were examined and scored for the number of vulva. In this way, we identified 10 genes as suppressors of the Muv phenotype in let-60(n1046gf), and 6 genes as enhancers (Figure 1A). We re-assayed these genes through three confirmatory rounds of screening and identified four consistent enhancers of the let-60(n1046gf) Muv phenotype (Figure 1B): dgk-2/F46H6.2, H37N21.1, mig-15/ZC504.4 and hpk-1/F20B6.8.
Figure 1.
C. elegans let-60 (n1046gf) RNAi kinase screen and mammalian in vitro transformation analysis. (A) Muv phenotype of let-60 (n1046gf) worms after RNAi-mediated knockdown of 656 worm kinase genes. The percentage of animals with a multivulva phenotype is shown. (B) We took the top 2% of genes and performed a second round of screening to identify four robust enhancers of the Muv phenotype. (C) RNAi against H37N21.1 causes excessive LET-60 signalling in vulval cells. egl-17::cfp transgenic worms showing ectopic expression of CFP in P5.p descendants after one cell division (P5.px), which indicates excessive LET-60 signalling. Also shown is the normal CFP expression in the P6.p descendants (P6.px). We observed ectopic expression at a rate of 18% (n=23). Top image is light field and bottom image is CFP expression. (D) Histogram of colony numbers from transformants derived from NIH3T3 cells cotransfected with an Nrbp1 shRNA vector (1–4) and pBabe-RasV12 vector. pBabe-RasV12 vector alone (R) and empty pBabe vector (N) were used as controls. (E) Western blot analysis of Nrbp1 knockdown in NIH3T3 cells. (F) Histogram of transformants after BJ-ET-st p53kd, p16kd cells were transfected with NRBP1 human shRNA vectors (1–6) or pRS empty vector control (N). (G) BJ-ET-st p53kd, p16kd transformed colonies transfected with human NRBP1 shRNA vector 3 were picked, cultured and whole-cell lysates collected for use in pull-down assays using GST- Raf1-BSD. Lysates were separated and visualised using a pan-RAS antibody (L—whole cell lysate, GTP—positive control, GDP—negative control, N—untransfected control cell pull-down, 3—shRNA trasformant pull-down). Data are represented as mean±s.d.; *P<0.05, **P<0.01. All shRNA hairpin sequences are available in the Supplementary methods. ***P<0.001.
dgk-2 encodes a diacylglycerol (DAG) kinase, and cooperation between activating mutations in RAS and DAG-activated signalling pathways in mammalian transformation assays is well established (Castagna et al, 1982; Nakamura et al, 1989). We confirmed that dgk-2 loss enhances the let-60(n1046gf) Muv phenotype using a mutant (null) allele of dgk-2 (dgk-2(gk124)); worms homozygous for dgk-2(gk124) showed no obvious phenotype, whereas 100% of let-60(n1046gf); dgk-2(gk124) double-mutant worms showed the Muv phenotype due to hyperinduction of vuval progenitor cells (data not shown). In contrast, the mechanism of action for the other three genes was less clear; mig-15 encodes a Nick-interacting kinase homologue that is known to play a role in cell migration and has been implicated in integrin and WNT signalling (Poinat et al, 2002); hpk-1 encodes a homeodomain-interacting kinase homologue, and H37N21.1 encodes an orthologue of the adapter protein NRBP1.
gap-1 encodes a member of the Ras GTPase-activating protein family and negatively regulates the let-60 pathway with respect to vulval development. Worms homozygous for a gap-1 loss-of-function allele (gap-1(ga133)) show normal vulval induction (vulval index=3.0) (Hajnal et al, 1997); however, loss of negative regulators of Ras signalling such as lip-1 (Berset et al, 2001) can yield a Muv phenotype in these mutants (vulval index>3.0). RNAi targeting of H37N21.1 in gap-1(ga133) mutants resulted in 17% of worms displaying the Muv phenotype (vulval index of 3.7); however, no vulval defects were observed when H37N21.1 was targeted by RNAi in wild-type worms (vulval index of 3.0). This phenotype is indicative of a hyperactivated LET-60 signalling cascade. To further implicate LET-60 signalling, we used a more sensitive assay (egl-17::cfp) that reports on the levels of LET-60 signalling, without the gap-1 mutation in the background, and directly in the vulval cells. Normally, only the descendants of P6.p will express CFP because it receives the highest amount of EGF (LIN-3), but not P5.p or P7.p descendants. We found that worms depleted of H37N21.1 ectopically express CFP and are therefore displaying excessive LET-60 signalling (Figure 1C). Thus we focused our efforts on H37N21.1. H37N21.1 is an orthologue of NRBP1 and shows 57.3% amino-acid sequence identity. NRBP1 and Nrbp1 from mouse show 99.6% amino-acid sequence identity (Supplementary Figure 1).
Knockdown of NRBP1 cooperates with activated Ras to induce transformation
To determine if NRBP1 interacts with the RAS pathway in mammals, we performed focus formation assays in murine NIH3T3 fibroblasts. Cells stably transfected with Nrbp1 shRNA did not form foci. In contrast, cells transfected with pBabe-RasV12 showed a modest number of transformed foci; however, transfection of pBabe-RasV12 into cell lines expressing shRNAs against Nrbp1 resulted in a significant number of foci (in 3/4 Nrbp1 shRNAs assayed; Figure 1D). Knockdown of Nrbp1 protein was confirmed by western blotting (Figure 1E). In agreement with these findings, shRNA-mediated stable knockdown of NRBP1 in the BJ-ET-st p53kd, p16kd human fibroblast cell line, which readily transforms following subtle perturbation of the RAS pathway or the pathways that regulate the RAS signalling cascade (Voorhoeve and Agami, 2003), resulted in a significant increase in the number of transformed colonies in soft agar (in 5/6 NRBP1 shRNAs assayed; Figure 1F). However, stable repression of NRBP1 saw no increase in the activation of RAS as assayed by western blotting following pull-down using GST-Raf1-BSD (Figure 1G). Thus, just as knockdown of H37N21.1 in C. elegans cooperates with gain-of-function mutations of let-60, knockdown of NRBP1 cooperates with, but does not directly activate, RAS in the transformation of mammalian cells in culture. To follow this further, we examined the effect of Nrbp1 loss in vivo.
Generation and analysis of Nrbp1 conditional and knockout mice
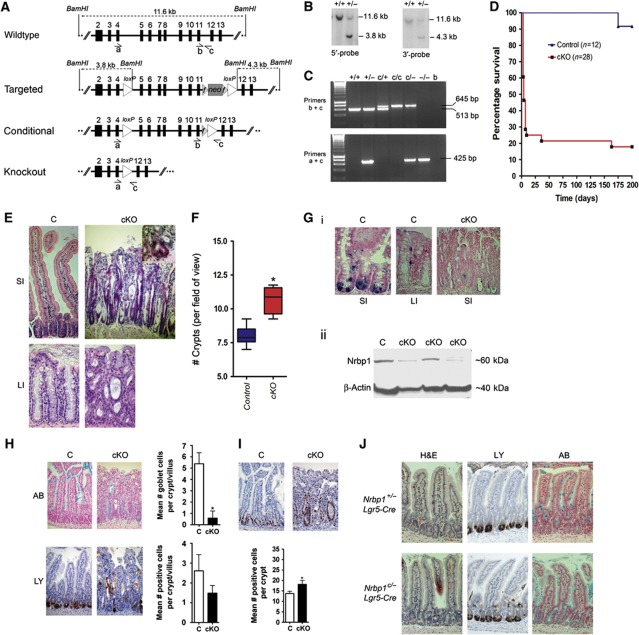
To examine the effect of loss of Nrbp1 in vivo, we created a conditional knockout mouse model. The targeting vector consisted of 9.6 kb of homology to the Nrbp1 locus, with a loxP site inserted in intron 4, and a flrted PGK–neomycin selection cassette and second loxP site inserted in intron 11 (Figure 2A). Correctly targeted E14J ES cells were identified by Southern blot analysis (Figure 2B). Knockout (Nrbp1+/−) and conditional (Nrbp1+/flox) Nrbp1 alleles were established in mice by breeding male chimeras with Cre and Flp deleters, respectively (Figure 2C). Excision of exons 5–11 disrupts the kinase-like domain of Nrbp1 and we hypothesised that this would result in nonsense-mediated decay of the remaining transcript.
Figure 2.
Generation and characterisation of Nrbp1 knockout mice. (A) Schematic representation of the Nrbp1 locus, along with targeted, conditional and knockout Nrbp1 alleles. The PGK–neomycin selection cassette (neo) is flanked by Frt sites (striped triangles). Open triangles indicate LoxP sites for conditional deletion of exons 5–11. (B) Confirmation of correct targeting was performed by Southern blot analysis of BamHI-digested DNA; The wildtype band for both 5′- and 3′-probes was 11.6 kb and the targeted band was 3.8 and 4.3 kb, respectively. (C) Subsequent genotyping was performed by PCR, using either primers ‘b/c’ to detect the wild-type and conditional allele, or primers ‘a/c’ to detect the knock out allele. (D) Survival curve of Nrbp1flox, RosaCreERT2/+ (cKO) mice and controls after intraperitoneal dosing with 3 mg tamoxifen per day for 3 days. (E) H&E staining of the SI (small intestine) and LI (large intestine) from C (control; Nrbp1+/−RosaCreERT2/+ or Nrbp1flox/+RosaCreERT2/+) and cKO (Nrbp1flox/floxRosaCreERT2/+ or Nrbp1flox/−RosaCreERT2/+) mice at 5–7 days post tamoxifen treatment showing crypt elongation by primitive looking cells ( × 200 magnification). Inset: cKO intestines show evidence of crypt fission. (F) cKO mouse intestines had more crypts per field of view than C intestines at 4–5 days post dosing. (G) (i) ISH analysis using probe specific to Nrbp1 in the SI (small intestine) and LI (large intestine) of C mice (control; × 400 magnification) and the SI of cKO mice ( × 200 magnification) at 5 days post tamoxifen treatment, showing reduction of Nrbp1 expression levels in cKO tissue. (G) (ii) Western blot analysis of intestinal tissue harvested from C and cKO mice at 5 days post treatment with 1 mg tamoxifen for 4 days shows a reduction of Nrbp1 protein levels. β-Actin levels used as a control. (H) Histological and immunohistological analysis showing cKO mouse intestines had fewer goblet cells (as determined by Alcian blue staining) and an altered distribution of Paneth cells (as determined by lysozyme staining) compared to C mice at 4–5 days post dosing. *P<0.05. (I) cKO mice had more proliferating cells (as determined by Ki67 immunopositivity) than C intestines at 4–5 days post dosing. Data are represented as mean±s.d.; *P<0.05. (J) Histological and immunohistochemical analysis of the small intestines of 7–8-week-old Nrbp1−/cLgr5-EGFP-IRES-CreERT2+ and Nrbp1−/+Lgr5-EGFP-IRES-CreERT2+ control mice at 4–5 post dosing with 1 mg tamoxifen for 3 days. Data are represented as mean +/− s.d. (n=6 per genotype). Stem cell-specific deletion of Nrbp1 resulted in abnormal paneth cell localisation and granulisation, and abnormal goblet cell production ( × 200 magnification; haematoxylin and eosin, H&E; anti-lysozyme antibody, LY; Alcian blue, AB). **P<0.01, ***P<0.001.
Mice heterozygous for the knockout Nrbp1 allele (Nrbp1+/−) were grossly normal at birth and fertile (data not shown); however, homozygosity resulted in embryonic lethality at day E7.5 (Supplementary Figure 2). Therefore, loss of Nrbp1 is not essential for cell viability (since null embryos are able to implant) but defects in or after gastrulation appear to result in embryonic lethality.
Perturbation of proliferation and differentiation of the intestinal epithelium following conditional loss of Nrbp1
To establish the phenotypic effects of somatic loss of Nrbp1, conditional Nrbp1 mice (Nrbp1flox/flox and Nrbp1flox/−) were intercrossed with mice carrying the RosaCreERT2 allele (RosaCreERT2/CreERT2), which express Cre-ERT2 protein ubiquitously, and at all developmental stages (Hameyer et al, 2007). At 7–8 weeks of age, Nrbp1flox/floxRosaCreERT2/+ or Nrbp1flox/−RosaCreERT2/+ mice (which showed no phenotypic differences from each other and hereafter are collectively referred to as cKO mice) and Nrbp1flox/floxRosa+/+, Nrbp1+/−RosaCreERT2/+ or Nrbp1flox/+RosaCreERT2/+ mice (hereafter collectively referred to as control mice) were dosed with tamoxifen (intraperitoneal injection of 4 mg tamoxifen for three consecutive days). cKO mice became moribund as early as 3 days after dosing (n=11/28) and 75% died or became moribund by day 9 (n=21/28, Figure 2D). In contrast, no control mice died within 174 days post treatment (Figure 2D; P<0.0001).
cKO mice showed macroscopically distended stomachs where food had failed to pass into the intestines and oedematous intestinal tracts with an abnormal mucosal appearance. Histopathological analysis of the brain, kidneys, skin, thymus, heart, pancreas, lungs, uterus/testis and spleen of control and cKO mice revealed no abnormalities. However, cKO mice showed zone 3 hydropic changes in their livers, indicative of malnutrition (data not shown), and the intestines showed a complete loss of architecture, including reduced numbers of differentiated cells, cells showing dysplastic nuclear atypia, widespread crypt elongation by primitive looking cells and reduced villous length (Figure 2E). An increased level of crypt fission was also observed (Figure 2F). To examine the expression pattern of Nrbp1 in the intestine, we performed in situ hybridisation (ISH) analysis using a probe specific to the conditionally deleted region of Nrbp1. In control intestines, expression of Nrbp1 was found to be highest in the Paneth, enteroendocrine and precursor goblet cell lineages; however, Nrbp1 expression was also found in the crypt and to overlap with the Lgr5 and Olfm4-positive stem cells (Supplementary Figure 3). To confirm the specificity of the Nrbp1 in situ probe, we analysed tamoxifen-treated cKO tissues and as expected little or no Nrbp1 expression was observed (Figure 2G). Similarly, western blotting showed a downregulation in Nrbp1 protein levels in the intestine of tamoxifen-treated cKO mice (Figure 2G).
Histological analysis of each of the cell types that constitute the intestinal epithelium, revealed most notably a marked decrease in the number of Alcian Blue positive goblet cells, an altered distribution of Paneth cells (Figure 2H) and reduced enteroendocrine cells (Supplementary Figure 3). In addition, expression of the proliferation marker Ki67 and the number of mitotic figures were significantly increased, while expression of the apoptotic marker cleaved caspase 3 was not significantly changed (Figure 2I, Supplementary Figure 3). To further investigate the phenotypic effect of Nrbp1 deficiency specifically within the intestinal stem cell compartment, we utilised the Lgr5-EGFP-IRES-CreERT2 knock-in mouse, which expresses CreERT2 specifically within the Lgr5-positive stem cell compartment with recombination occurring in a sporadic and low-penetrant fashion (<10% intestinal stem cells) upon tamoxifen treatment (Barker et al, 2007; Barker et al, 2009). Immunohistochemical analysis of small intestines from Nrbp1−/cLgr5-EGFP-IRES-CreERT2+ mice at 5 days post tamoxifen treatment (intraperitoneal dosing of 1 mg/ml tamoxifen for 3 days) showed a similar phenotype to cKO mice, albeit to a lesser extent due to the very low level of recombination, with Paneth cells displaying abnormal localisation and granularisation, and abnormal goblet cell production (Figure 2J).
Together, these data indicate that loss of Nrbp1 protein in the adult intestines of mice causes aberrant proliferation of progenitor-like cells in the crypts and increased crypt fission, which profoundly affects the normal programmes of cellular differentiation and localisation along the crypt–villus axis. These features are similar to several mouse models engineered to perturb the WNT pathway (Batlle et al, 2002; Sansom et al, 2004; Madison et al, 2005; Finch et al, 2009).
Expression profiling of Nrbp1-deficient intestinal cells
To evaluate the role of loss of Nrbp1 in intestinal homoeostasis, we performed microarray expression profiling on small intestinal epithelial cells from tamoxifen-dosed control and cKO mice. For these experiments the Cre driver used was RosaCreERT2/+. The expression of 3331 genes was significantly altered (P<0.01 to 2 decimal places between these groups (Supplementary Table 1). Interestingly, a number of WNT-responsive genes showed significantly increased expression in cKO mice compared with controls, including Ccnd1, Cd44, Mmp7, Sox9 and Tnfrs12a.
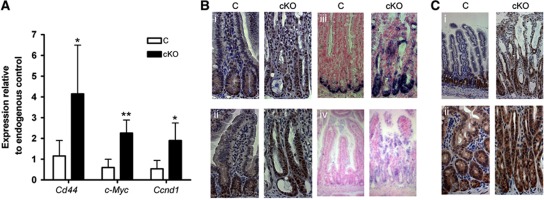
To validate the expression arrays, we examined the expression of a number of WNT-responsive genes by quantitative RT–PCR (qRT–PCR). Expression of Cd44, c-Myc and Ccnd1 were upregulated more than three-fold in the intestinal epithelium of cKO mice compared with controls (Figure 3A). Further examination of WNT target genes by immunohistochemistry and ISH analysis of the small intestines revealed increased staining for c-Myc, Sox9, Mmp7 and Lgr5 in cKO mice compared with controls (Figure 3B). In addition, cKO mice showed a widespread increase in pErk nuclear positivity and β-catenin staining in the mucosa that matched the upward spread of crypt progenitor-like cells (Figure 3C). In addition, although the clearest signature was for the WNT pathway, we note that a number of other signalling pathways were also significantly altered—for example, expression analysis of the Notch genes revealed that Notch3 and Notch4 were both significantly overexpressed and this in turn lead to an increase in Hes1 expression (Supplementary Figure 4).
Figure 3.
Expression analysis of conditional Nrbp1 mice. (A) qRT–PCR analysis of the WNT target genes, Cd44, c-Myc and Ccnd1 in the intestine of cKO and C mice at 4–5 days post tamoxifen administration. (B) Immunohistochemical (i, ii) and in situ analysis (iii, iv) of small intestines from C and cKO mice at 7–9 days post tamoxifen treatment indicating loss of Nrbp1 causes an increase in (i) c-Myc ( × 400 magnification), (ii) Sox9 ( × 400 magnification), (iii) Mmp7 ( × 400 magnification) and (iv) Lgr5 ( × 200 magnification). (C) Immunohistochemical analysis of small intestines from C and cKO mice at 7–9 days post tamoxifen treatment show increased expression of (i) pErk ( × 200 magnification), but no obvious increase in (ii) nuclear Ctnnb1 signal ( × 400 magnification). *P<0.05, **P<0.01.
We next compared our differentially expressed gene set to gene expression signatures of genetic and chemical perturbations using the Molecular signatures database MSigDB Version 2.5 ( http://www.broadinstitute.org/gsea/msigdb/index.jsp) (Subramanian et al, 2005). There was a striking overlap (P=5.89 e−23 and P=5.5 e−47) with genes downregulated at 4 or 5 days following conditional deletion of adenomatous polyposis coli (Apc) in the mouse intestine (Sansom et al, 2004) (Supplementary Figure 5). Similarly, there was a significant overlap between genes that were upregulated upon loss of either Apc or Nrbp1 (P=0.007 (5 days) P=6.2E−06 (4 days); Supplementary Figure 5), suggesting that similar changes to the programmes of gene expression occur following loss of either gene. These expression changes may also reflect changes in the constitution of the cells that make up the intestinal epithelium.
To further investigate the role of Nrbp1 within the intestine, we grew Nrbp1 cKO and control crypts ex vivo. Using adenoviral cre infection of organoid cultures to delete Nrbp1, we observed decreased organoid budding and organoid dis-organisation (Supplementary Figure 6). After long-term culture of these organoids they eventually regained the ability to bud, presumably due to the outgrowth of non-recombined cells (Koo et al, 2011). On removal of R-spondin from early cultures all crypts of all genotypes died, indicating that Nrbp1 null crypts are still dependant on Wnt signalling.
Nrbp1 interacts with Elongin BC E3 ubiquitin ligase complex
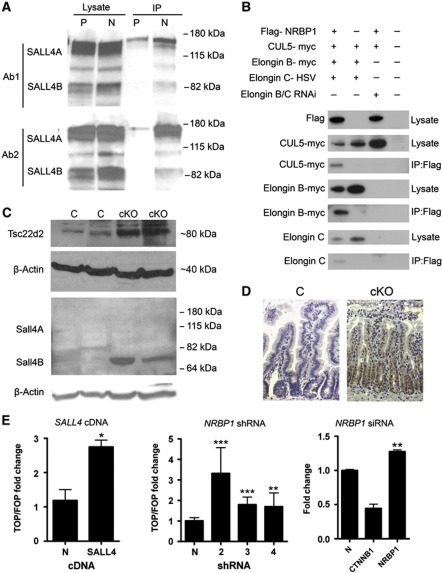
To further explore possible roles for Nrbp1 within mammalian cells, we performed tandem affinity purification followed by mass spectrometric analysis using a FTAP-tagged human NRBP1 cDNA expression plasmid in mouse ES cells (E14J; 129P2/Ola). Five proteins were shown to bind strongly to NRBP1, including Tceb1 (Elongin B), Tceb2 (Elongin C), Tsc22d2, Tsc22d4 and Sall4 (Supplementary Table 2). To validate these results, we performed single affinity purifications from human HEK293 cells expressing the same FTAP-tagged human NRBP1 cDNA. We were able to recover TCEB1, TCEB2, TSC22D2 and TSC22D4, among others. SALL4 was not recovered, most likely due to the stem cell-specific expression of SALL4; we therefore confirmed that Sall4 binds to Nrbp1 in mouse ES cells by coimmunoprecipitation (Figure 4A). We also confirmed binding of Nrbp1 to Elongin B and Elongin C by coimmunoprecipitation (Figure 4B). Since ELONGIN B and the TSC22s have been shown to bind to NRBP1, and NRBP1 carries both a functional Elongin BC-binding motif (BC-Box) and a CUL5-binding site (Supplementary Figure 1) (Rual et al, 2005; Mahrour et al, 2008; Gluderer et al, 2010), we hypothesised that it may function as a substrate recognition subunit of a Cul5 E3 ligase complex. Indeed, expression of NRBP1, ELONGIN B, ELONGIN C and CUL5 in HEK293 cells allowed coimmunoprecipitation of CUL5 with NRBP1, only when ELONGIN B and ELONGIN C were present in the lysate (Figure 4B). This is in contrast to a previous report suggesting that NRBP1 is not part of a complex with CUL5 (Mahrour et al, 2008). To further investigate if NRBP1 could be functioning within an Elongin BC E3 ubiquitin ligase complex, we performed western blot and immunohistochemical analysis on intestinal tissue from tamoxifen-treated cKO mice and found a significant increase in both Tsc22d2 and Sall4B protein levels compared to controls (Figures 4C and D and Supplementary Figure 7) suggesting that Nrbp1 may have a role in the turnover or accumulation of these proteins. It is, however, important to note that as yet we have been unable to functionally link Nrbp1 to the ubiquitination of these proteins. Intriguingly, overexpression of SALL4 has been shown to play a role in regulating the WNT pathway in the haematopoietic system through a direct physical interaction with β-catenin (Ma et al, 2006). Here, we show that knockdown of NRBP1 or overexpression of SALL4 in HCT-116 human colon cancer cells results in an upregulation of WNT reporter activity (Figure 4E) suggesting a direct link between NRBP1 and the WNT pathway through SALL4, an avenue we are currently investigating further.
Figure 4.
NRBP1 interacts with components of the Elongin BC E3 ubiquitin ligase complex. (A) Immunoprecipitation with anti-Flag antibody of mouse ES cells carrying either Flag-tagged NRBP1 cDNA expression plasmid (N) or Flag-tagged Phactr2 cDNA expression plasmid (P) as a control, followed by western blotting with one of two different anti-Sall4 antibodies (Ab1 and Ab2), showed coprecipitation of both Sall4 isoforms with Nrbp1 and not the control. In mouse ES cells, Sall4 has been shown to run at 85 and 140 kDa. (B) Immunoprecipitation of CUL5 and other members of the ubiquitination machinery with NRBP1 from HEK293 cells. (C) Western blot analysis with anti-Tsc22d2 antibody, anti-Sall4 and anti-β-Actin control antibody of intestinal tissue harvested at day 5 from cKO (Nrbp1floxRosaCreERT2/+) or C (control; Nrbp1floxRosa+/+) mice treated with 1 mg tamoxifen for 4 days. In somatic cells, Sall4 has been shown to run at 66 and 113 kDa. (D) Immunohistochemical analysis of Sall4 in the intestinal tissue from C (control; Nrbp1floxRosa+/+) and cKO (Nrbp1floxRosaCreERT2/+) mice at 4–5 days post tamoxifen treatment showing elevated levels of Sall4. (E) Transcriptional activation of TCF/LEF measured using the dual luciferase assay (TOP/FOP) in HCT-116 colon cancer cells transfected with a Sall4 cDNA plasmid (left panel) or an NRBP1 shRNA construct (middle panel). N refers to an empty vector control. Transcriptional activation of TCF/LEF was also measured by siRNA-mediated knockdown of NRBP1 in SW180 WNT reporter cells (right panel). Knockdown of CTNNB1 was used as a control. *P<0.05, **P<0.01, ***P<0.001.
Nrbp1 is a tumour suppressor in vivo
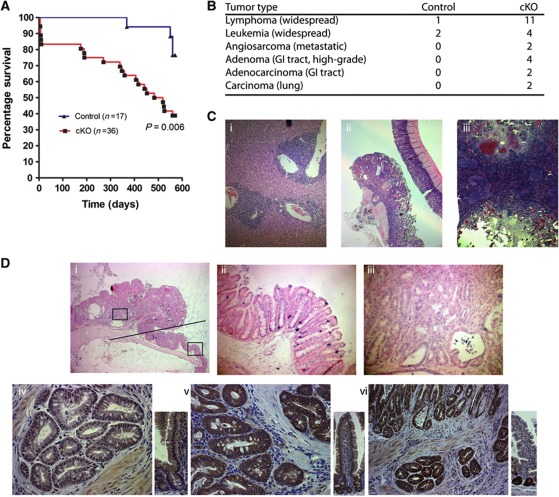
Given Nrbp1 proficiency is vital for intestinal homoeostasis and loss of Nrbp1 cooperates with oncogenic Ras and can modulate Wnt signalling, we hypothesised that Nrbp1 may be a tumour suppressor in vivo. In our initial survival analysis (Figure 2D), 18% (n=5) of cKO mice survived the acute intestinal phenotype associated with loss of Nrbp1, and histological analysis of their intestines revealed no abnormalities. This suggested incomplete recombination of the Nrbp1 allele and intestinal regeneration by the surviving cells (Muncan et al, 2006). To generate a cohort of mice for tumour watch, we treated 7–8-week-old control and cKO mice with a lower dose of tamoxifen (intraperitoneal dose of 1 mg tamoxifen for four consecutive days) and placed them on tumour watch. cKO mice showed significantly reduced survival compared to controls, which was associated with an increased incidence of tumourigenesis, including lymphomas/leukaemias and solid tumours (P=0.006; Figure 5A–C). Interestingly, 38% (6/16) of the cKO mice displayed gastrointestinal tract tumours, specifically four were adenomas with high-grade dysplasia and two were invasive adenocarcinoma (5/6 tumours were located in the caecum; Figure 5B). In situ analysis of these tumours confirmed loss or reduction in Nrbp1 RNA levels (Figure 5Di), and immunohistochemical analysis showed widespread nuclear c-Myc and pErk staining (Figure 5Dii–iv), analogous to the staining patterns identified in the ‘early Nrbp1-null phenotype’. Taken together, these data are all consistent with a role for Nrbp1 as a tumour suppressor gene in vivo.
Figure 5.
Nrbp1 is a tumour suppressor in vivo. (A) Survival analysis of cKO (NrbpfloxRosaCreERT2/+) and control mice (Nrbpflox/+RosaCreERT2/+ and Nrbpflox/floxRosa+/+) after treatment with 1 mg tamoxifen per day for 4 days. Animals became moribund and were culled as a result of either an ‘early Nrbp1 loss phenotype’ (animals <10 days post treatment) or when they developed macro/microscopically visible tumour(s) (animals >10 days post treatment). (B) Table provides a summary of the tumour types found in the mice (some animals had more than one type of tumour). (C) H&E staining of tissues from cKO mice showing (i) widespread lymphoma in the liver ( × 100 magnification), (ii) invasive colorectal adenocarcinoma ( × 250 magnification) and (iii) high-grade angiosarcoma in the spleen ( × 250 magnification). (D) ISH analysis of Nrbp1 in a colorectal lesion from an Nrbp1floxRosaCreERT2/+ mouse shows (i) a tumour area left of the black line, and normal colonic tissue right of the black line ( × 25 magnification), with the lower box enlarged in (ii) and the upper box enlarged in (iii), showing reduced Nrbp1 expression levels in tumour tissue ( × 200 magnification). Immunohistochemical analysis of an invasive colorectal adenocarcinoma from an Nrbp1floxRosaCreERT2/+ mouse showing significant strongly positive staining with (iv) c-Myc ( × 400 magnification) and (vi) pErk ( × 200 magnification) antibodies, but (v) no increased nuclear Ctnnb1 signal ( × 400 magnification).
Reduction in Nrbp1 expression is seen in a wide variety of human tumours and correlates with poor prognosis
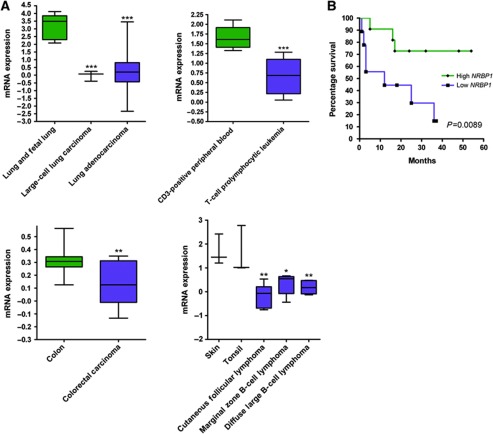
To further explore the association of NRBP1 loss in cancer, we performed expression analysis on a wide variety of human tumour types (including leukaemia, lymphoma, colorectal, breast, brain, oesophageal, renal cell, prostatic and lung cancers) and found significantly reduced NRBP1 expression levels in many of these tumour types when compared with relevant control tissues (P<0.01; Figure 6A). As lung adenocarcinoma showed the most significant difference in NRBP1 expression levels between normal and tumour tissues, and survival analysis data was available, we examined the relationship between NRBP1 expression and survival and found that low NRBP1 expression strongly correlated with a poor prognosis (P=0.008; Figure 6B).
Figure 6.
Associations of altered NRBP1 expression with human cancers. (A) Expression analysis of NRBP1 in human large-cell lung carcinoma, lung adenocarcinoma, T-cell prolymphocytic leukaemia, colorectal carcinoma, cutaneous follicular lymphoma, marginal zone B-cell lymphoma and diffuse large B-cell lymphoma compared to relevant normal tissues. *P<0.05, **P<0.01 and ***P<0.001. (B) Survival analysis of patients with lung adenocarcinoma expressing either high (green) or low (blue) NRBP1 expression levels.
Discussion
Cancer is a multistep process that requires extensive synergy between oncogene activation and tumour suppressor gene loss. Many oncogenes are key components of highly conserved signalling pathways, and genetic screens in model organisms including Drosophila and C. elegans have identified oncogene orthologues in the EGF, WNT, NOTCH and TGFβ pathways, among others. However, such screens have yielded few orthologues of tumour suppressor genes. In this paper, we employed the power of RNAi screening in C. elegans to identify candidate tumour suppressor genes by screening for genes that enhanced developmental defects due to gain-of-function mutations in Ras. Importantly, we found that loss of H37N21.1, the worm orthologue of NRBP1, enhanced the effect of gain-of-function mutations in Ras suggesting that it may act as a tumour suppressor gene in mammals.
NRBP1 is a highly conserved adaptor protein that we show is required for early mouse embryonic development. In mammalian in vitro systems, we have shown that together with oncogenic RAS, NRBP1 knockdown leads to an increase in transformation; however, NRBP1 loss does not appear to affect RAS signalling directly suggesting that other pathways must modulate RAS activity following NRBP1 loss. To investigate the function of Nrbp1 in vivo, we constructed a conditional knockout mouse model. Two somatic phenotypes were discerned depending on the pattern of Nrbp1 disruption. First, there was an ‘early intestinal phenotype’ at 4–9 days post tamoxifen dosing. This phenotype consisted of expansion of crypt progenitor-like cells with aberrant proliferation in elongated crypts and increased crypt fission, together with abnormal differentiation and localisation of cells along the crypt–villus axis. Ultimately this phenotype led to abnormal intestinal function, malnutrition and the death of over 80% mice by 5–10 days (Figure 2). The intestinal epithelia of these mice showed several similarities to mouse models engineered to perturb the WNT pathway (Batlle et al, 2002; Sansom et al, 2004; Madison et al, 2005; Finch et al, 2009). Interestingly, the phenotype also shows a striking similarity to a model in which the POU-family transcription factor Oct4 was overexpressed in the intestine (Hochedlinger et al, 2005). We tentatively speculate that Nrbp1 regulates Sall4 levels through its role in an Elongin BC E3 ubiquitin ligase complex and thus suggest that the intestinal phenotype of the Nrbp1 cKO mice may possibly be due to increased levels of Sall4 influencing cell fate by perturbing the WNT pathway, which tips the balance of progenitor cells from differentiation to proliferation. As yet, however, we have been unable to conclusively prove this functional link. Furthermore, although we were able to pull-down complexes containing components of the ubiquitination machinery and Nrbp1, we have been unable to conclusively prove a role for Nrbp1 in protein turnover. Indeed, the data we have collected on Nrbp1 suggest that it may have pleiotropic effects on several pathways.
Most of the mice which survived this ‘early intestinal phenotype’ went on to develop a wide range of tumours, including intestinal adenomas in 38% of mice, a third of which had advanced to invasive intestinal adenocarcinoma, as well as lung adenocarcinomas, lymphomas, leukaemias and angiosarcomas, with significantly reduced survival. Thus, Nrbp1 acts as a tumour suppressor gene in vivo with Nrbp1 loss resulting in a tendency to intestinal adenoma/adenocarcinoma and lymphoma/leukaemia formation (Figure 5). Analysis of human cancer expression datasets showed that NRBP1 is downregulated in a range of tumour types, and that in lung adenocarcinomas low NRBP1 expression is associated with a poorer prognosis (Figure 6). It is intriguing to note that although we were able to recapitulate the acute intestinal phenotype associated with loss of Nrbp1 by expressing CreERT2 specifically in Lgr5+ stem cells (Figure 2J), and that loss of Nrbp1 activates the WNT pathway (Figure 3), we have not observed intestinal polyposis or tumourigenesis in Nrbp1−/cLgr5-EGFP-IRES-CreERT2+ mice up to 100 days (data not shown). This may be because of the modest activation of the WNT pathway we observed and because fewer cells underwent recombination using the Lgr5-EGFP-IRES-CreERT2 allele. In both models of Cre recombination and in ex vivo crypt culture, it appears that selection against Nrbp1 loss occurs suggesting that by itself Nrbp1 is unlikely to be an initiating event in colorectal cancer, instead cooperating mutations are likely to be required for tumourigenesis.
We have shown that NRBP1 interacts with ELONGIN B and C and CUL5, which are components of an E3 ubiquitin ligase complex. The Elongin BC complex is a heterodimer composed of ELONGIN C and ELONGIN B, that was initially identified as a positive regulator of RNA polymerase II elongation factor ELONGIN A (Reines et al, 1999). A more recognised role of Elongin BC is within the tumour suppressor complex together with the von Hippel-Lindau (VHL) protein, which targets the transcription factor hypoxia inducible factor-1α for ubiquitination (Duan et al, 1995; Kibel et al, 1995). Interaction of the Elongin BC complex with proteins such as VHL and suppressors of cytokine signalling (SOCS) depends on binding to a ∼10 amino-acid degenerate sequence motif, referred to as the BC-box, which has the consensus sequence of [(A/P/S/T)-L-X3-(C/S/A)-X3-(A/I/L/V)], a motif also present in NRBP1 (Supplementary Figure 1). The BC-box-containing protein mediates substrate binding and specificity, while Elongin BC functions as an adapter that links it to a Cullin/Rbx module that functions to recruit and activate an E2 ubiquitin conjugating enzyme for ubiquitylation of substrates. In addition to VHL and SOCS, a number of novel BC-box proteins have been shown to assemble with Cullin/Rbx and therefore it has been predicted that a larger family of ELONGIN BC-based E3 ubiquitin ligases exists (Kamura et al, 2001). We therefore suggest that NRBP1 is involved in substrate recognition within a RING-type E3–ligase complex to enhance the ubiquitin-mediated degradation of proteins. As yet, however, confirming this hypothesis has proved elusive. Potential substrates of NRBP1 could include TSC22d2 and SALL4, which we show bind strongly to NRBP1 and both show increased protein levels in vivo when Nrbp1 is deleted in the mouse (Figure 4). Sall4 is a transcription factor that plays an essential role in maintaining embryonic stem cell pluripotency and self-renewal (Warren et al, 2007). Interestingly, overexpression of SALL4 has been shown to cause aberrant activation of the WNT pathway in myelodysplastic syndromes (Shuai et al, 2009), and SALL4 expression has recently been linked to gastric cancers (Ushiku et al, 2010). Furthermore, sem-4, the C. elegans orthologue of SALL4, promotes vulval cell fate determination. TSC22d2 is a member of the TSC22 (TGFβ-stimulated clone 22) family of putative transcription factors that is activated by TGFβ1 receptor signalling (Jay et al, 1996). Tsc22 has been shown to be expressed at sites of epithelial mesenchymal interactions from the early stages of mouse development (Dohrmann et al, 1999). Indeed when we performed ISH analysis of intestinal tissue from Nrbp1 cKO mice, Tsc22d1 and Tsc22d4 were both strongly expressed in the pericryptal fibroblasts while Tsc22d2 was expressed primarily in the crypt regions (Supplementary Figure 6) suggesting a possible role in signalling from the intestinal mesenchyme to cryptal cells. The TSC22 protein family is conserved between humans and C. elegans, but the function of these proteins has been difficult to determine due to differences in isoforms, redundancy, synergistic and/or antagonistic functions (Gluderer et al, 2010). Much of the work on these proteins has been carried out in Drosophila, and the data have suggested genetic interactions with the fly orthologues of BMP, WNT, HH, NOTCH and components of the EGFR signalling pathway (Treisman et al, 1995; Dobens et al, 2000; Dobens et al, 2005). Furthermore, in this system the fly orthologue of NRBP1 (Madm) and the long isoform of TSC22, Bunched (bunA), have been shown to directly bind, and loss of either protein leads to similar growth phenotypes. It is interesting to note that TSC22D1 has been shown to be highly induced by oncogenic RAS expression and together with TSC22D4 regulate oncogene-induced senescence (Homig-Holzel et al, 2011).
In conclusion, using a novel combination of RNAi screens in worms together with detailed follow-up experiments in mouse and human cells, we show that NRBP1 is a tumour suppressor that plays a critical role in intestinal cell homoeostasis. Crucially, NRBP1 levels are reduced in a wide variety of human tumours and, furthermore, reduced NRBP1 levels in lung cancer correlates with a poor prognosis.
Materials and methods
C. elegans RNAi
Nematode strains used in this work were provided by the Caenorhabditis Genetics Centre. Screening was performed at 16 °C in 12-well plates using 715 bacterial RNAi clones (Supplementary Table 3) to target 656 transcripts with kinase or kinase-like motifs from the Ahringer library (Kamath et al, 2003). Bacterial RNAi feeding strains were grown overnight in 2TY plus 100 μg/ml ampicillin at 37 °C and spotted onto plates containing LB, 100 μg/ml ampicillin and 4 mM IPTG. Approximately 10 L4 staged worms were dispensed to the first well of each row and grown for 72 h, after which 1 adult worm was moved to a separate well and left overnight before removal. Embryos were left to mature for at least 72 h and scored for Muv in 100–200 animals in triplicate by visual inspection using a dissecting microscope. Muv was scored in comparison to the phenotype of the worm strain alone. The top and bottom 2% of genes were retested in two separate confirmatory rounds of screening. For egl-17::cfp assays, L3 animals were mounted and examined for persistent expression of egl-17::cfp prior to L4.
Cell culture, transformation assays and luciferase assays
All cell lines (NIH3T3, HEK 293, BJ-ET-st p53kd,p16kd, SW480 and HCT-116) were cultured in DMEM supplemented with 10% fetal bovine serum and glutamine/penicillin/streptomycin. For focus formation assays, NIH3T3 cells were transfected using lipofectamine (Invitrogen) with 200 ng of pBabe-RasV12 plasmid DNA together with 200 ng of pRS shRNA plasmid DNA designed against Nrbp1 (the sequence of shRNA hairpins is provided in the Supplementary methods) or an empty vector control. Cells were seeded at 1 × 105 per 6-cm plate and fed every 4 days for 2 weeks and then stained with methylene blue and foci were counted. Each experiment involved five replicate transfections performed on three separate occasions. BJ-ET-st p53kd,p16kd soft agar assays were performed as described previously (Voorhoeve and Agami, 2003) with human pRS shRNAs (provided in Supplementary methods), which were modified to introduce a blasticidin resistance cassette driven by the PGK promoter. Data were statistically analysed using the Mann–Whitney U-test. Luciferase assays were performed in 24-well plates with 7.5 × 103 HCT-116 or HEK 293 cells. Cells were cotransfected using lipofectamine with either 200 ng NRBP1 shRNA vectors or empty vector in combination with either 150 ng M50 Super 8x TOPFlash or M51 Super 8 × FOPFlash and 50 ng Renilla (pRL-SV40) plasmid. At 72 h after transfection, cells were lysed in 100-μl passive lysis buffer, and the dual luciferase assay performed according to the manufacturers protocol (Promega). The relative luciferase activity was calculated against Renilla activity. Each shRNA was assayed in experiments involving six replicates and these experiments were performed on at least two separate occasions. Data were statistically analysed using ANOVA. CellSensor assays were performed in 96-well plates with 2.4 × 104 SW480 cells. Cells were transiently transfected with 20 nm siRNA (Dharmacon) using Lipofectamine RNAimax (Invitrogen). At 72 h after transfection FRET read-out was measured using beta-lactamase loading solution (Invitrogen) by standard procedures. Data were statistically analysed using ANOVA. Ex vivo crypts were isolated from Nrbp cKO and control mice small intestine by incubating in 2 mM EDTA in PBS. Crypt cultures were grown in Matrigel (BD biosciences) and provided with Advanced DMEM/F23 (Invitrogen) supplemented with N2 (Invitrogen), B27 (Invitrogen), EGF (Peprotech), NOGGIN (Peprotech) and mR-Spondin (R&D Systems) (Sato et al, 2009). Adenoviral infection of crypts was performed by mechanically separating crypts away from Matrigel and incubating with Adeno-Cre or Adeno-GFP (as a control) virus at 37 °C for 3–4 h before re-plating in Matrigel. GFP expression was seen in ∼80% of cells the day after infection.
Mice
To generate the Nrbp1 targeting vector, DNA fragments for the 5′- and 3′-homology arms were amplified from bMQ-76E6 BAC (Adams et al, 2005) by PCR using the following primers: Capture arms1F—AAAAAAAAAAGAGCTCCTGGTCCTGTAGTTTTAGATTCCATG, Capture arms1R—TTT TTTTTTTGAATTCAAAGCGATACCCTGGAAAGTAAACT, Capture arms2F—AAAAAAAAAAGAATTCACGGGCACTGGGGAAGCTGAGTTCTA, Capture arms2R—TTTTTTTTTTGGTACCCATCTGCAAGGTAGGGGGAGGGGCGG, and cloned into pBS SK(+). The linear gap repair plasmid was transformed into EL350 cells carrying the bMQ-76E6 BAC (Liu et al, 2003) and gap-repaired plasmids were selected for ampicillin resistance. A LoxP site was inserted into intron 4 and a PGK–neomycin selection cassette flanked by Frt sites and a second LoxP site were introduced into intron 11 of the captured Nrbp1 DNA fragment. Targeting was performed in E14J (129P2/Ola) ES cells and germline chimeras were transmitted onto a C57BL/6J background. Southern blot analysis of BamHI-digested DNA was performed using 5′- and 3′-probes using standard procedures (5′-ProbeF—GAGGCTTTCTGGGTTTCCTC, 5′-ProbeR—CGGGGCACTTAGAGAAAAGA, 3′-ProbeF—GTTCGGGACTACTGGTGAGC and 3′-ProbeR—TGGCAATCAGCAATAAGACG). Mice were genotyped by PCR using the primers: A—GAA AAG GTC CGT GCA GTG TT, B—AGA TGG CAG TGC TGG AGA TT and C—CAA GCG TTC TTG TTC AGA CG. Male Nrbp1 mice carrying the unmodified targeted allele of Nrbp1 were crossed with either CMV-Cre (Su et al, 2002) or 129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J (FLPeR) mice (Farley et al, 2000) to produce Nrbp1+/− and Nrbp1flox/+ mice, respectively. RosaCreERT2/+ mice (Vooijs et al, 2001) were used for somatic deletion of Nrbp1, and Cre-mediated site-specific recombination was performed in mice aged 7–8 weeks carrying the RosaCreERT2/+ allele by intraperitoneal injection with either 100 μl of 40 mg/ml tamoxifen base in sunflower oil for three consecutive days or 100 μl of 10 mg/ml tamoxifen base in sunflower oil for four consecutive days. Mice that became moribund were euthanized, and tissues collected and fixed in 10% neutral buffed formalin. H&E staining was performed using standard procedures. All procedures with animals were carried out in accordance with Home Office UK guidelines
Immunohistochemistry, ISH and immunoblotting
Immunostaining was performed on 4-μm sections using the DAKO Autostainer Plus with the rabbit VECTASTAIN ELITE ABC horseradish peroxidase kit (Vector Laboratories). For primary antibodies, see Supplementary methods. In all cases, a total of 30 crypts/villi were counted from the small intestine of four Nrbp1flox/floxRosaCreERT2/+ and control mice. Four lengths of intestinal epithelium were photographed and used to estimate crypt numbers of Nrbp1flox/floxRosaCreERT2/+, Nrbp1flox/floxRosaCreERT2/+ and Nrbp1flox/+RosaCreERT2/+ mice (four of each genotype). All comparisons were made using the Mann–Whitney U-test. Western blotting was performed using standard methods. For primary antibodies see Supplementary methods. Activation of Ras was analysed using Active Ras Pull-Down Detection Kit, which contains GST–Raf1–RBD fusion protein (Thermo scientific) in accordance with the manufacturer’s instructions. ISH was performed with 6-μm sections on the automated Ventana Discovery platform with RiboMap and BlueMap Kits (Ventana) and DIG-labelled riboprobes (primers for generating probes are available in the Supplementary methods). Immunopreciptiations were performed with either HEK293 cells or mouse embryonic stem cells using a FLAG Immunoprecipitation Kit (Sigma). HEK293 cells were transiently trasfected with the following expression plasmids: NRBP1-Flag-MSCV, pcDNA3-myc-CUL5 (Ohta et al, 1999) and c-Myc-Elongin B and HSV-Elongin C (Brower et al, 1999). Elongin B and Elongin C siRNAs pools were purchased from Thermo scientific. ES cells were stably integrated with NRBP1-Flag-MSCV after selection with neomycin.
qRT–PCR and microarray analysis
Total RNA was isolated from snap-frozen small intestine using TRIzol (Invitrogen), DNase treated using Turbo DNase (Ambion) and 2 μg total RNA reverse transcribed using the BD Sprint kit with random hexamers (ABI) according to the manufacturer’s protocols. qRT–PCRs were performed with SYBR Green (ABI) on the ABI StepOnePlus Real-Time PCR Systems in accordance with the manufacturer’s instructions (see Supplementary methods for primer sequences). The final quantitation was determined using the standard curve method relative to the house-keeping genes Gapdh or Actb. Data were statistically analysed using ANOVA. Microarray analysis was performed on total RNA using the Illumina mouse-6 expression beadchip version 2 platform (Illumina) by standard procedures. Probe summaries were calculated in BeadStudio and quantile normalised. A linear model fit was applied and the top differentially expressed genes were tabulated for each contrast using the Benjamini and Hochberg method to correct the P-values. The Molecular signatures database MSigDB Version 2.5 ( http://www.broadinstitute.org/gsea/msigdb/index.jsp) was used to compute overlaps between the top and bottom 500 most differentially expressed genes to gene sets in MSigDB (Subramanian et al, 2005).
Tandem affinity purification mass spectrometry analysis
The protocol used for the purification of proteins has been described previously (Pardo et al, 2010). Briefly, HEK293 and ES cells expressing the CBP2TEV3FLAG tag were lysed and incubated with anti-FLAG M2 Dynal beads (Sigma). TEV eluates from mouse ES cells were subsequently incubated with calmodulin resin (Stratagene), from which protein complexes were eluted by Ca chelation with EGTA. For single affinity purifications from HEK293 cells, the second purification step was omitted. EGTA and TEV eluates were separated by one-dimensional SDS electrophoresis. Gel lanes were excised, digested with trypsin and peptides extracted with 0.5% formic acid–50% acetonitrile. Peptides were analysed with on-line nanoLC-MS/MS on a LTQ FT mass spectrometer (ThermoElectron) coupled with an Ultimate 3000 Nano/Capillary HPLC System (Dionex). The LC used a PepMap C18 trap (0.3 mm id × 5 mm, LC Packings) and a BEH C18 analytical column (75 μm id × 10 cm, Waters) with a 30 min linear gradient of 4–32% CH3CN/0.1% formic acid. The LTQ FT was operated in standard data dependent acquisition. The survey scans (m/z 400–2000) were acquired on the FT-ICR at a resolution of 100 000 at m/z 400. The top four multiply charged ions were subject to MS/MS in the linear ion trap at an isolation width of 2 Th. Dynamic exclusion width was set at ±20 p.p.m. for 45 s. The automatic gain control target value was regulated at 5E5 for FT and 1E4 for the ion trap, with maximum injection time at 1000 ms for FT and 200 ms for ion trap, respectively. The raw files were processed with BioWorks (Thermo). Database searches were performed with Mascot v.2.2 (Matrix Science) against the Human or mouse IPI database (v. December 2008). The search parameters were: Trypsin/P with two missed cleavages, 10 p.p.m. mass tolerance for MS, 0.5-Da tolerance for MS/MS, fixed modification carbamidomethyl (C), and variable modifications of Acetyl (Protein N-term), Deamidated (NQ), Dioxidation (M), Formyl (N-term), Gln->pyro-Glu (N-term Q), Methyl (E) and Oxidation (M). Decoy database searches were performed at the same time as the real searches, resulting in false discovery rates under 5%.
Supplementary Material
Acknowledgments
Work in the DJ Adams laboratory and in the MJ Arends laboratory is funded by Cancer Research UK (CR-UK) and the Wellcome Trust. Work in the AG Fraser laboratory is funded by CCS. LvdW is supported by a Kay Kendall Leukaemia Fund fellowship. GBP is funded by an MRC Career Development Award (G0600127). GP is a Pfizer Fellow of the Life Sciences Research Foundation. We thank Team 17, 77 and 83 of the Wellcome Trust Sanger Institute for their technical support and to Owen Sansom for helping us establish the organoid culture system.
Author contributions: CHW, CC, LvdW, GP, MP, TG, LY, JC, GBP, REM, DJW and NHM performed the experiments. AGR performed the computational analysis. MJA performed the histopathological analysis. MJA, AGF and DJA led the project. CHW, LvdW and DJA wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A (2005) A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758 [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007 [DOI] [PubMed] [Google Scholar]
- Batlle E, Henderson JT, Beghtel H, van den Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering M, Pawson T, Clevers H (2002) Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell 111: 251–263 [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Clark SG, Horvitz HR (1990) Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348: 503–509 [DOI] [PubMed] [Google Scholar]
- Berset T, Hoier EF, Battu G, Canevascini S, Hajnal A (2001) Notch inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291: 1055–1058 [DOI] [PubMed] [Google Scholar]
- Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR (2006) Emerging roles of pseudokinases. Trends Cell Biol 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Brower CS, Shilatifard A, Mather T, Kamura T, Takagi Y, Haque D, Treharne A, Foundling SI, Conaway JW, Conaway RC (1999) The elongin B ubiquitin homology domain. Identification of Elongin B sequences important for interaction with Elongin C. J Biol Chem 274: 13629–13636 [DOI] [PubMed] [Google Scholar]
- Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y (1982) Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem 257: 7847–7851 [PubMed] [Google Scholar]
- Dobens L, Jaeger A, Peterson JS, Raftery LA (2005) Bunched sets a boundary for Notch signaling to pattern anterior eggshell structures during Drosophila oogenesis. Dev Biol 287: 425–437 [DOI] [PubMed] [Google Scholar]
- Dobens LL, Peterson JS, Treisman J, Raftery LA (2000) Drosophila bunched integrates opposing DPP and EGF signals to set the operculum boundary. Development 127: 745–754 [DOI] [PubMed] [Google Scholar]
- Dohrmann CE, Belaoussoff M, Raftery LA (1999) Dynamic expression of TSC-22 at sites of epithelial-mesenchymal interactions during mouse development. Mech Dev 84: 147–151 [DOI] [PubMed] [Google Scholar]
- Duan DR, Pause A, Burgess WH, Aso T, Chen DY, Garrett KP, Conaway RC, Conaway JW, Linehan WM, Klausner RD (1995) Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269: 1402–1406 [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM (2000) Widespread recombinase expression using FLPeR (flipper) mice. Genesis 28: 106–110 [PubMed] [Google Scholar]
- Finch AJ, Soucek L, Junttila MR, Swigart LB, Evan GI (2009) Acute overexpression of Myc in intestinal epithelium recapitulates some but not all the changes elicited by Wnt/beta-catenin pathway activation. Mol Cell Biol 29: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Gluderer S, Brunner E, Germann M, Jovaisaite V, Li C, Rentsch CA, Hafen E, Stocker H (2010) Madm (Mlf1 adapter molecule) cooperates with Bunched A to promote growth in Drosophila. J Biol 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A, Whitfield CW, Kim SK (1997) Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev 11: 2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 modulates microRNA activity in Caenorhabditis elegans. Cell 136: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameyer D, Loonstra A, Eshkind L, Schmitt S, Antunes C, Groen A, Bindels E, Jonkers J, Krimpenfort P, Meuwissen R, Rijswijk L, Bex A, Berns A, Bockamp E (2007) Toxicity of ligand-dependent Cre recombinases and generation of a conditional Cre deleter mouse allowing mosaic recombination in peripheral tissues. Physiol Genomics 31: 32–41 [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R (2005) Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121: 465–477 [DOI] [PubMed] [Google Scholar]
- Homig-Holzel C, van Doorn R, Vogel C, Germann M, Cecchini MG, Verdegaal E, Peeper DS (2011) Antagonistic TSC22D1 variants control BRAF(E600)-induced senescence. EMBO J 30: 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper JD, Baker E, Ogbourne SM, Sutherland GR, Antalis TM (2000) Cloning of the cDNA and localization of the gene encoding human NRBP, a ubiquitously expressed, multidomain putative adapter protein. Genomics 66: 113–118 [DOI] [PubMed] [Google Scholar]
- Jay P, Ji JW, Marsollier C, Taviaux S, Berge-Lefranc JL, Berta P (1996) Cloning of the human homologue of the TGF beta-stimulated clone 22 gene. Biochem Biophys Res Commun 222: 821–826 [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237 [DOI] [PubMed] [Google Scholar]
- Kamura T, Burian D, Yan Q, Schmidt SL, Lane WS, Querido E, Branton PE, Shilatifard A, Conaway RC, Conaway JW (2001) Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J Biol Chem 276: 29748–29753 [DOI] [PubMed] [Google Scholar]
- Kibel A, Iliopoulos O, DeCaprio JA, Kaelin WG Jr. (1995) Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 269: 1444–1446 [DOI] [PubMed] [Google Scholar]
- Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, van Es JH, Clevers H (2011) Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods 9: 81–83 [DOI] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG (2003) A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res 13: 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Cui W, Yang J, Qu J, Di C, Amin HM, Lai R, Ritz J, Krause DS, Chai L (2006) SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 108: 2726–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison BB, Braunstein K, Kuizon E, Portman K, Qiao XT, Gumucio DL (2005) Epithelial hedgehog signals pattern the intestinal crypt-villus axis. Development 132: 279–289 [DOI] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW (2008) Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J Biol Chem 283: 8005–8013 [DOI] [PubMed] [Google Scholar]
- Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H, Clarke AR (2006) Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf-4 target gene c-Myc. Mol Cell Biol 26: 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Kishi Y, Pajares MA, Rando RR (1989) Structural basis of protein kinase C activation by tumor promoters. Proc Natl Acad Sci USA 86: 9672–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell 3: 535–541 [DOI] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J (2010) An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell 6: 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinat P, De Arcangelis A, Sookhareea S, Zhu X, Hedgecock EM, Labouesse M, Georges-Labouesse E (2002) A conserved interaction between beta1 integrin/PAT-3 and Nck-interacting kinase/MIG-15 that mediates commissural axon navigation in C. elegans. Curr Biol 12: 622–631 [DOI] [PubMed] [Google Scholar]
- Reines D, Conaway RC, Conaway JW (1999) Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr Opin Cell Biol 11: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, Batlle E, Simon-Assmann P, Clevers H, Nathke IS, Clarke AR, Winton DJ (2004) Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev 18: 1385–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459: 262–265 [DOI] [PubMed] [Google Scholar]
- Shuai X, Zhou D, Shen T, Wu Y, Zhang J, Wang X, Li Q (2009) Overexpression of the novel oncogene SALL4 and activation of the Wnt/beta-catenin pathway in myelodysplastic syndromes. Cancer Genet Cytogenet 194: 119–124 [DOI] [PubMed] [Google Scholar]
- Solari F, Ahringer J (2000) NURD-complex genes antagonise Ras-induced vulval development in Caenorhabditis elegans. Curr Biol 10: 223–226 [DOI] [PubMed] [Google Scholar]
- Su H, Mills AA, Wang X, Bradley A (2002) A targeted X-linked CMV-Cre line. Genesis 32: 187–188 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Lai ZC, Rubin GM (1995) Shortsighted acts in the decapentaplegic pathway in Drosophila eye development and has homology to a mouse TGF-beta-responsive gene. Development 121: 2835–2845 [DOI] [PubMed] [Google Scholar]
- Ushiku T, Shinozaki A, Shibahara J, Iwasaki Y, Tateishi Y, Funata N, Fukayama M (2010) SALL4 represents fetal gut differentiation of gastric cancer, and is diagnostically useful in distinguishing hepatoid gastric carcinoma from hepatocellular carcinoma. Am J Surg Pathol 34: 533–540 [DOI] [PubMed] [Google Scholar]
- Vooijs M, Jonkers J, Berns A (2001) A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep 2: 292–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhoeve PM, Agami R (2003) The tumor-suppressive functions of the human INK4A locus. Cancer Cell 4: 311–319 [DOI] [PubMed] [Google Scholar]
- Warren M, Wang W, Spiden S, Chen-Murchie D, Tannahill D, Steel KP, Bradley A (2007) A Sall4 mutant mouse model useful for studying the role of Sall4 in early embryonic development and organogenesis. Genesis 45: 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.