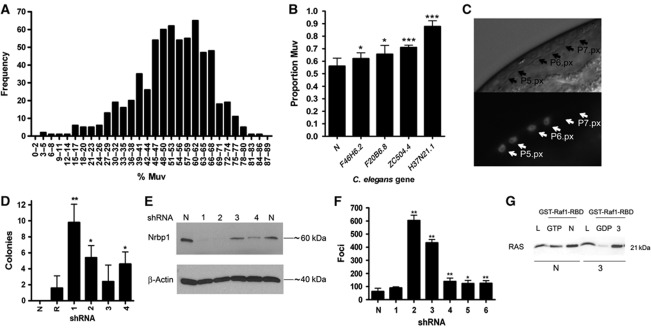
Figure 1.
C. elegans let-60 (n1046gf) RNAi kinase screen and mammalian in vitro transformation analysis. (A) Muv phenotype of let-60 (n1046gf) worms after RNAi-mediated knockdown of 656 worm kinase genes. The percentage of animals with a multivulva phenotype is shown. (B) We took the top 2% of genes and performed a second round of screening to identify four robust enhancers of the Muv phenotype. (C) RNAi against H37N21.1 causes excessive LET-60 signalling in vulval cells. egl-17::cfp transgenic worms showing ectopic expression of CFP in P5.p descendants after one cell division (P5.px), which indicates excessive LET-60 signalling. Also shown is the normal CFP expression in the P6.p descendants (P6.px). We observed ectopic expression at a rate of 18% (n=23). Top image is light field and bottom image is CFP expression. (D) Histogram of colony numbers from transformants derived from NIH3T3 cells cotransfected with an Nrbp1 shRNA vector (1–4) and pBabe-RasV12 vector. pBabe-RasV12 vector alone (R) and empty pBabe vector (N) were used as controls. (E) Western blot analysis of Nrbp1 knockdown in NIH3T3 cells. (F) Histogram of transformants after BJ-ET-st p53kd, p16kd cells were transfected with NRBP1 human shRNA vectors (1–6) or pRS empty vector control (N). (G) BJ-ET-st p53kd, p16kd transformed colonies transfected with human NRBP1 shRNA vector 3 were picked, cultured and whole-cell lysates collected for use in pull-down assays using GST- Raf1-BSD. Lysates were separated and visualised using a pan-RAS antibody (L—whole cell lysate, GTP—positive control, GDP—negative control, N—untransfected control cell pull-down, 3—shRNA trasformant pull-down). Data are represented as mean±s.d.; *P<0.05, **P<0.01. All shRNA hairpin sequences are available in the Supplementary methods. ***P<0.001.