Figure 6. AGEs suppressed gene expression of PPARγ and reduced its trans-activity in HSCs, which were diminished by curcumin.
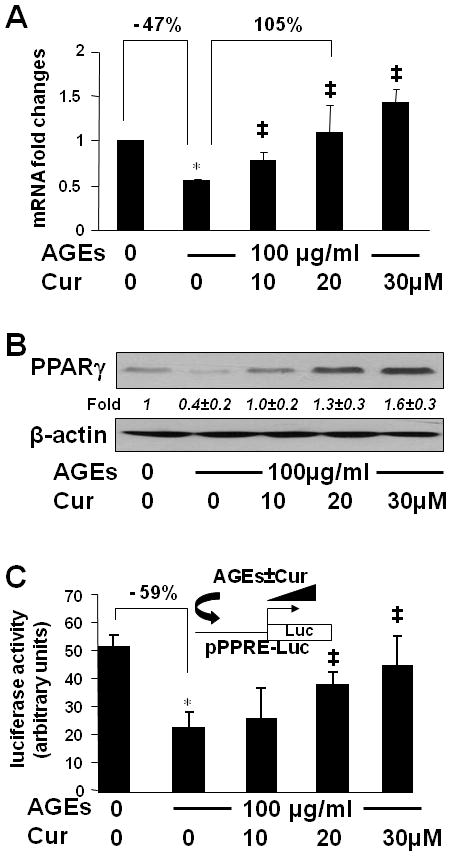
(A & B) Serum-starved HSCs were stimulated with or without AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media for 24 hr. Total RNA and whole cell extracts were prepared. (A). real-time PCR assays. Values were presented as mRNA fold changes (mean ± s. d., n=3). *p<0.05 vs. cells with no treatment (the 1st column). ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). (B) Western blotting analyses. β-actin was used as an internal control for equal loading. Representatives were from three independent experiments. Italic numbers beneath blots were fold changes (mean ± s. d., n=3) in the densities of the bands compared with the control without treatment in the blot, after normalization with the internal invariable control.
(C) HSCs were transfected with the PPARγ activity luciferase reporter plasmid pPPRE-Luc. After recovery, cells were serum-starved for 4 hr prior to the treatment with or without AGEs (100 μg/ml) in the presence of curcumin (0–30 μM) in serum-depleted media with PGJ2 (5 μM) for 24 hr. Luciferase activity assays were conducted (n=6). *p<0.05 vs. cells with no treatment (the 1st column), ‡p<0.05 vs. cells treated with AGEs alone (the 2nd column). The floating schema denoted the plasmid pPPRE-Luc in use for transfection and the application of AGEs w/wt curcumin to the system.
