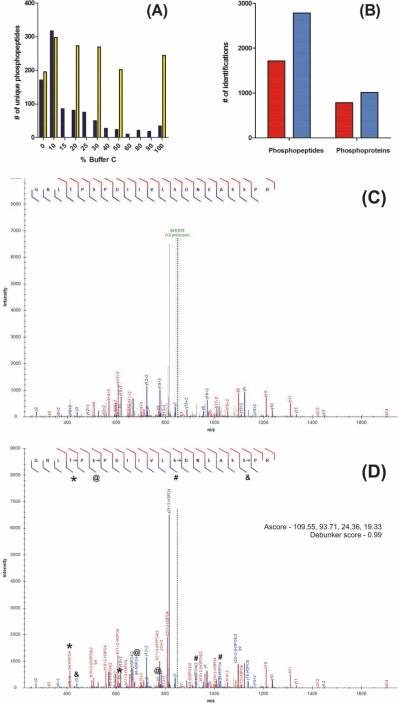
Figure 1.
MudPIT optimization of HAP-enriched phosphopeptides. (A) 500 μg of a whole cell tryptic digest was enriched using a 250 μm × 5 cm in-house packed HAP column. Enriched peptides were transferred to a MudPIT column and analyzed using either a 12 hr 6-step MudPIT (yellow bars) or 24 hr 12-step MudPIT (blue bars). (B) The number of phosphopeptides and phosphoproteins identified with (dark blue bars) and without (red) the MSA search option in ProLuCID. Annotated spectra for the quadruply modified phosphopeptides R.GRLpTPpSPDIIVLpSDNEASpSPR.S with (D) and without (E) the ProLuCID MSA search option.