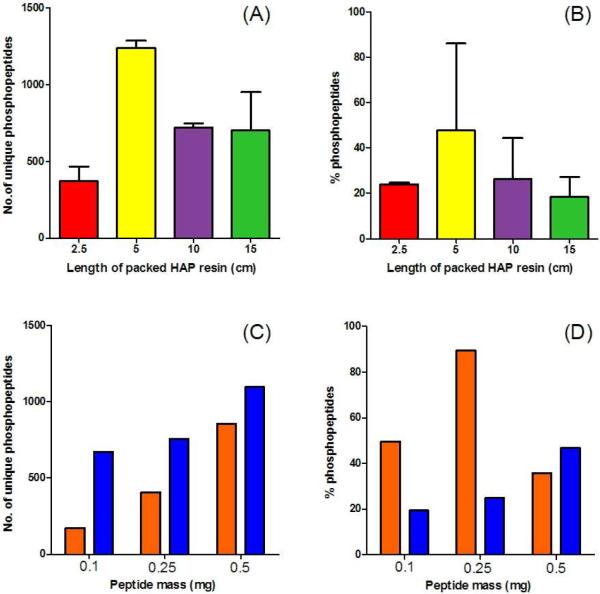
Figure 2.
HAP enrichment optimization. Duplicate phosphopeptide enrichments of 500 μg of a whole cell tryptic digest were performed using different 250 μm in-house packed HAP columns of 2.5, 5, 10, and 15 cm in length corresponding to 1.25, 2.5, 5, and 7.5 mg HAP resin, respectively. Enriched peptides were transferred to a MudPIT column and analyzed using a 6-step MudPIT for 12 hrs with a LTQ-Orbitrap XL. The number of unique phosphopeptides identified (A) and the % of peptides identified as phosphopeptides (B) are plotted for these experiments. Single phosphopeptide enrichments of different masses of tryptic digests (100, 250, and 500 μg) were performed using individual 250 μm in-house packed HAP columns. Based on the optimization in (A), for each 100 μg of lysate to be enriched 1 cm (0.5 mg) of HAP resin was packed into the microcolumn. Enriched peptides were transferred to a MudPIT column and analyzed using a 6- step MudPIT for 12 hrs using either a LTQ-Orbitrap XL (orange bars) or LTQ-Orbitrap Velos (blue bars). The number of unique phosphopeptides identified (C) and the % of peptides identified as phosphopeptides (D) are plotted for these experiments.