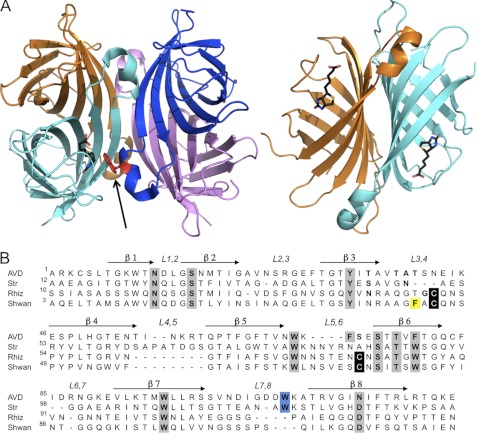
FIGURE 1.
Structures of tetrameric avidins. A, tetrameric assembly of egg white avidin (left), with respective monomers shown in magenta, blue, cyan, and orange. The biotin ligand is shown in black in the binding site of the cyan monomer (for clarity). The Trp residue (shown in red) from an adjacent monomer (blue) contributes to the high affinity of biotin binding and is indicated by an arrow. The 1-4 sandwich-like monomer-monomer interaction (right) forms numerous stabilizing interactions (orange and cyan). All molecular graphics figures were generated using PyMOL (47). B, multiple sequence alignment of avidin (AVD), streptavidin (Str), rhizavidin (Rhiz), and shwanavidin (Shwan). The eight β-strands forming the tertiary structure are indicated by arrows, and the loops connecting the strands are labeled. Residues participating in biotin binding are highlighted in gray, cysteines participating in the disulfide bridge in the new binding site are in black, and the Trp residues from the L7,8 contributing to the biotin binding in tetrameric avidins are highlighted in blue. Phe-43 from the L3,4 loop compensates for the lack of the intermonomeric Trp residue and is highlighted in yellow. Shwanavidin exhibits 46, 50, and 61% sequence similarity and 26, 33, and 47% identity compared with avidin, streptavidin, and rhizavidin, respectively.