Fig. 7.
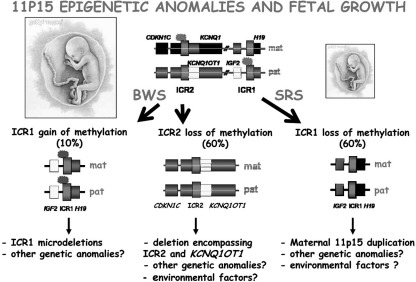
The two imprinted domains of the 11p15 chromosomal region are under the control of two ICRs. The reciprocal imprinting of the maternally (mat) expressed H19 and the paternally (pat) expressed IGF2 depends on an ICR1 located upstream from the H19 gene that acts as an insulator. The repressor factor CTCF (CCCTC-binding factor) binds to the unmethylated maternal copy of the ICR and prevents the IGF2 gene promoter from interacting with enhancers downstream from the H19 gene. This results in transcriptional silencing of the maternal IGF2 allele. On the paternal allele, the ICR is methylated, and CTCF binding is prevented. This leads to IGF2 transcription on the paternal allele and silencing of the H19 gene. The centromeric KCNQ1 domain produces a noncoding RNA (antisense KCNQ1OT1 RNA) that silences many of the genes in this domain. Paternally expressed genes are represented as white boxes, maternally expressed genes as black boxes, and nonexpressed genes as gray boxes. BWS is associated with a variety of genetic and epigenetic defects within the imprinted 11p15 region. Most patients (70%) exhibit an epigenetic defect. Ten percent of BWS patients display an imprinting defect at the IGF2-H19 domain (aberrant GOM at the maternal copy of the ICR), which results in silencing of the maternal H19 gene and a biallelic expression of the IGF2 gene. The majority of the BWS patients exhibit a LOM at the ICR of the KCNQ1 domain. Loss of methylation at this ICR results in activation of the normally silent maternal allele of KCNQ1OT1 and CDKN1C silencing. In SRS, the mirror phenotype of BWS, a loss of imprinting at the IGF2–H19 domain was identified: the paternal allele switches to a maternal epigenotype, and this results in biallelic expression of H19 and loss of IGF2 expression. Genetic and environmental factors could induce these epigenetic anomalies.