Abstract
Thymocytes expressing the NKT cell semi-invariant αβ TCR are thought to undergo agonist interactions with CD1d ligands prior to expressing PLZF, a BTB-POZ transcription factor that directs acquisition of the effector program of these innate-like T cells. Whether PLZF can mediate this effector conversion independently of agonist signaling has not been investigated. We demonstrated that transgenic expression of PLZF under the CD4 promoter induced the innate effector program in two different MHC class II-restricted TCR transgenic Rag1−/− models examined. In CD4 thymocytes expressing a fixed transgenic TCR β chain, the associated TCR α sequences in wild-type and PLZF-transgenic mice overlapped extensively, further demonstrating that PLZF could induce the effector program in most CD4 T cells that would normally be selected as naÔve cells. In contrast, PLZF altered the negative selection of thymocytes expressing TCR β chains reactive against several retroviral superantigens. Thus, PLZF is the first example of a transcription factor inducing an effector program in the absence of T cell agonist interactions or cell division. Its expression may also enhance the survival of agonist-signaled thymocytes.
Introduction
The development of lymphocytes as naÔve cells that express a vast diversity of antigen receptors and traffic to lymph nodes is central to adaptive immunity. Upon recognition of antigen in this environment, lymphocytes typically undergo clonal expansion, acquisition of specialized effector functions and migration to infected tissues. In contrast with this well-established scenario, unconventional subsets of B and T cells undergo expansion and effector differentiation during development, following exposure to endogenous rather than foreign ligands (1). These innate-like lymphocytes express distinctive patterns of differentiation and tissue homing depending on the expression of canonical BCRs and TCRs, thereby affording early protection against different types and routes of microbial infections. The cellular and molecular mechanisms governing the development of NKT cells (2–4) and other unconventional lymphocyte subsets, including recently identified innate lymphocytes (5, 6), is a matter of considerable biological and evolutionary interest.
Recent studies have demonstrated that three distinct populations of innate-like T cells, CD1d-restricted Vα14(huVα24)-Jα18 NKT cells, MR1-restricted Vα19(huVα7.2)-Jα33 MAIT cells and Vγ1-Vδ6 NKT cells (whose ligand is unknown) expressed the signature transcription factor promyelocytic leukemia zinc finger PLZF (Zbtb16) (7–9). Mice deficient in PLZF lacked NKT lineage cells and instead redirected their progenitors to the conventional naÔve CD4 lineage (7, 8). In transgenic mice ectopically expressing PLZF under the cd4 or the lck promoter, an increased polyclonal population of CD44hiCD62Llow T cells with dual IL-4/IFN-γ production was observed (7, 10, 11). These striking findings from loss and gain of function experiments suggested the key role of PLZF in inducing the quintessential innate-like effector properties of NKT cells. Since NKT cells are autoreactive and thought to be selected by agonist TCR-ligand interactions, it is unclear whether agonist interactions are also required to induce the PLZF-mediated effector program in conventional T cells, or whether PLZF can act as a direct switch inducing the effector program independently of agonist signaling. While a recent report suggested that PLZF transgenic cells did not exhibit changes in staining with a panel of Vβ and Vα specific antibodies, the impact of PLZF expression on positive and negative selection of antigen-specific TCR αβ heterodimers has not been investigated.
Here, we examined the influence of PLZF expression on established models of TCR transgenic RAG−/− T cell development and on the selection of TCR alpha chains in thymocytes expressing a fixed transgenic Vβ chain. We concluded that ectopic expression of PLZF induced most CD4 thymocytes that would normally be selected as naÔve cells to become effector cells. Therefore PLZF appears to be the first transcription factor capable of inducing the effector T cell program in the absence of agonist signaling and cell division. In contrast, we found changes in the Vβ repertoire associated with recognition of several retrovirally-encoded superantigens, indicating that negative selection was significantly impaired by PLZF expression.
Materials and Methods
Mice
C57BL/6, BALB/c, Rag1−/− (B6.129S7-Rag1tm1Mom/J) and OTII (C57BL/6-Tg(TcraTcrb)425Cbn/J) mice were from The Jackson Laboratory. B6.129S2-H2-Ab1tm1Gru was from Taconic. PLZF-transgenic (7), Marilyn TCR-transgenic (12), and Rag2p-GFP reporter (13) mice were all in the C57BL/6 background.
For the generation of Vβ7 transgenic mice, Vβ7 TCRβ rearrangement was PCR amplified from cDNA prepared from a Vβ7+ NKT cell hybridoma 414.A2 (14) and cloned into the pTβcass plasmid (15). The linearized construct was injected into fertilized C57BL/6 oocytes, and the injected oocytes were implanted into pseudopregnant CD-1 (VAF+) outbred female mice. Transgenic mice were screened by flow cytometry and PCR (forward primer: 5'-gccttgtggacatgaaagt-3'; reverse primer: 5'-gtcagctttgagccttcacc-3').
All mice were raised in a specific pathogen-free environment at the University of Chicago, and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Flow Cytometry
Fluorochrome-labeled monoclonal antibodies (clone indicated in parentheses) against mouse CD3ε (17A2), CD4 (GK1.5 or L3T4), CD8α (53-6.7), CD24 (M1/69), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), TCRβ (H57-597), Vα2 (B20.1), V®5.1/5.2 (MR9-4), Vβ7 (tr310), V®11 (RR3-15), V®12 (MR11-1) and V®14 (14-2) were purchased from eBioscience, BD Biosciences, or BioLegend.
Samples were analyzed on an LSRII (Becton Dickinson), or sorted on a FACSAria (Becton Dickinson), with doublet exclusion and DAPI staining of dead cells.
TCR sequencing
For TCR sequencing, the following procedure was adapted from published work (16, 17). Vβ7-transgenic thymocytes were enriched for mature T cells using MACS and further sorted for Vα2+ thymocytes. cDNA was prepared from equivalent amounts of RNA derived of 50,000–500,000 cells using Oligo-d(T), as described above. cDNA was PCR amplified for 30 cycles using pan-Vα2 (TRAV14) (5’-agcagcaggtgagacaaa-3’) and Cα (5’-tcagtgatgaacgttccagattccatg-3’) primers, both derived of sequences available at IMGT (http://imgt.org), gel purified, and cloned into TOPO vector (Invitrogen). Products were transformed into XL-10 Gold ultracompetent cells (Stratagene) and plated for single colonies. Single colonies were picked, DNA-prepped, and sequenced with the M13Rev primer using standard procedures by the University of Chicago DNA Sequencing Facility. Sequences were submitted to IMGT VQUEST (http://imgt.org/IMGT_vquest/share/textes/) for alignment to known V and J genes, and mapping of P and N nucleotides. The resulting data set was filtered to remove sequences which had a “V-Region Identity %” of less than 98%, a “J-Region Identity %” of less than 95%, contained nonsense “AA Junction” translations, or were unable to be mapped by VQUEST. The filtered data set was organized and tallied using Microsoft Excel. Percent overlap was calculated as the mean of reciprocal comparisons between two populations.
in vitro T cell stimulation cultures
For CD69 induction, thymocytes were depleted of CD69+ cells using FITC-conjugated anti-CD69 and anti-FITC magnetic microbeads followed by AutoMACS cell separator (Miltenyi Biotec). 50,000 CD69-depleted thymocytes were plated with 400,000 CD3-depleted CD45.1+ splenocytes at a density of 2×106 cell/ml. Duplicate wells with OTII peptide at 0, 0.2, 1, 5, and 25 mg/ml were cultured for 20 hours at 37°C prior to analysis by flow cytometry.
Statistics
The unpaired, two-tailed, t-test was used for all statistical calculations.
Results
PLZF induces the effector program in CD4+ SP thymocytes prior to emigration
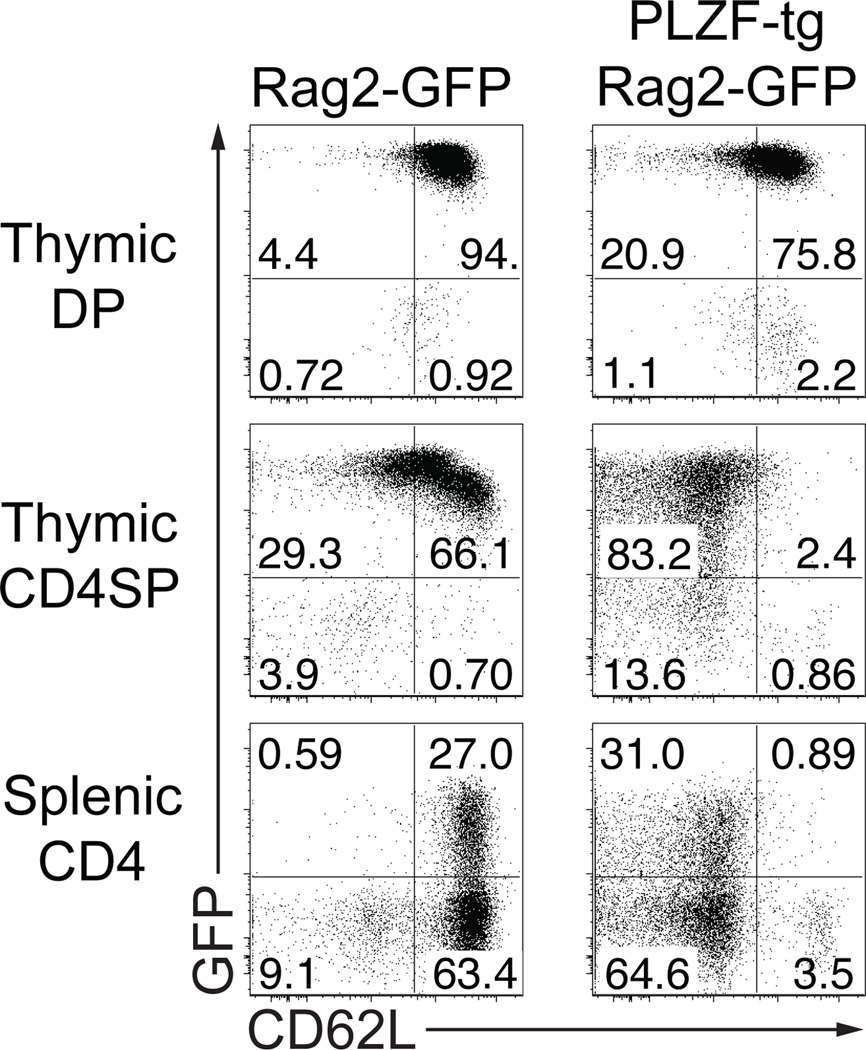
Transgenic expression of PLZF driven by the cd4 or Lck promoter induced notable changes in T cells that were typical of the NKT program (7, 10, 11), including the conversion from a naÔve lymph node trafficking (CD44loCD62Lhi) IL2-only producer state into an effector state capable of producing both IL4 and IFNγ and displaying tissue-rather than lymph node-homing properties (CD44hiCD62Llo). As the extent of this effector conversion was dependent on the dose of PLZF (10), we used the previously described CD4p-PLZF transgenic line #1797 where PLZF is expressed at levels matching those of developing NKT cells at stage 1 and 2 (corresponding to effector conversion). In these mice, >95% CD4 T cells acquire the effector phenotype (7). Previous studies showed that acquisition of the effector phenotype by PLZF-transgenic thymocytes and splenocytes did not seem to involve cell division based on BrdU or EdU incorporation studies (7, 11). Therefore, we used the Rag2p-GFP reporter mouse system (13) where, in the absence of cell division, green fluorescence decreases as a sole function of time after the DP stage and can be monitored to track recent thymic emigrants in the spleen (18). As shown in Fig. 1 (left column), expression of GFP reached the highest level in wild-type DP thymocytes, declined in CD4 SP thymocytes and was still detected on 27% of CD4 splenocytes representing recent thymic emigrants. Nearly all of these recent thymic emigrants expressed high levels of CD62L, as expected for conventional CD4 T cells. In contrast, while PLZF-transgenic CD4 cells (Fig. 1, right column) had a GFP profile similar to WT in thymus and spleen, >95% of them expressed the CD62Llow phenotype, whether in the thymus, in the spleen or among the recent thymic emigrants. These results further support the conclusion that PLZF expression can permanently convert most CD4 T cells into effector-type cells in the absence of cell division in the thymus before they emigrate to the spleen.
Figure 1. PLZF-transgenic CD4 SP thymocytes permanently downregulate CD62L prior to emigration.
PLZF-transgenic mice were bred to Rag2p-GFP-transgenic mice and the progeny was analyzed at 4–7 weeks of age. The plots show expression of GFP and CD62L among CD4+CD8+ (DP) and CD4+CD8− (CD4SP) thymocytes or TCRβ+CD4+ (CD4) splenocytes. Data are representative of three experiments.
PLZF-transgenic CD4 effector T cells are selected by MHC class II ligands expressed on thymic epithelial cells
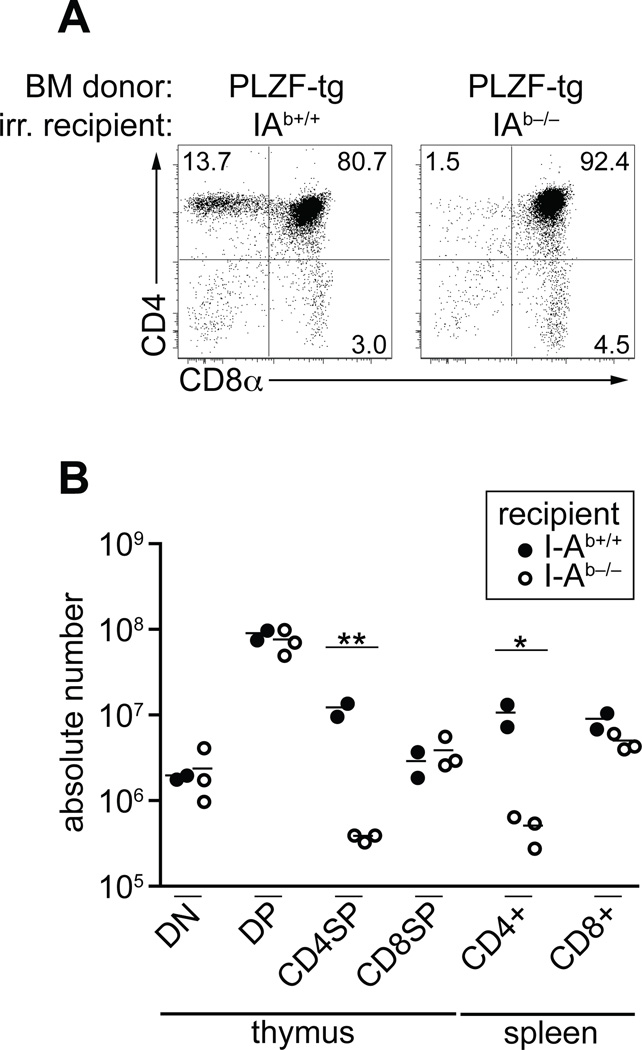
To test whether this conversion involved ‘conventional’ thymocytes recognizing MHC class II ligands displayed by epithelial cells, we generated PLZF-Tg into I-Ab−/− bone marrow radiation chimeras and compared them with PLZF-Tg into WT chimeras. Fig. 2A–B clearly showed that the selection of PLZF-Tg CD4 cells required MHC class II expression by host radioresistant cells as CD4 cells were severely reduced in the thymus and spleen of host mice lacking MHC class II.
Figure 2. PLZF-transgenic CD4 cells are selected by epithelial MHC II ligands.
PLZF-transgenic bone marrow was transferred into lethally irradiated I-Ab+/+ or I-Ab−/− recipients. (A) representative data from the thymus at 7 weeks post-transfer. (B) summary of T cell subset numbers in the thymus and spleen of individual chimeras. DN: CD4−CD8−, DP: CD4+CD8+. A similar requirement for epithelial MHC ligand was observed with another set of bone marrow chimeras where PLZF-transgenic bone marrow was injected into MHC-I and II deficient mice (β2M−/−IAb−/−). *p<0.05, **p<0.01.
PLZF expression converts conventional TCR-transgenic CD4 thymocytes into effector cells
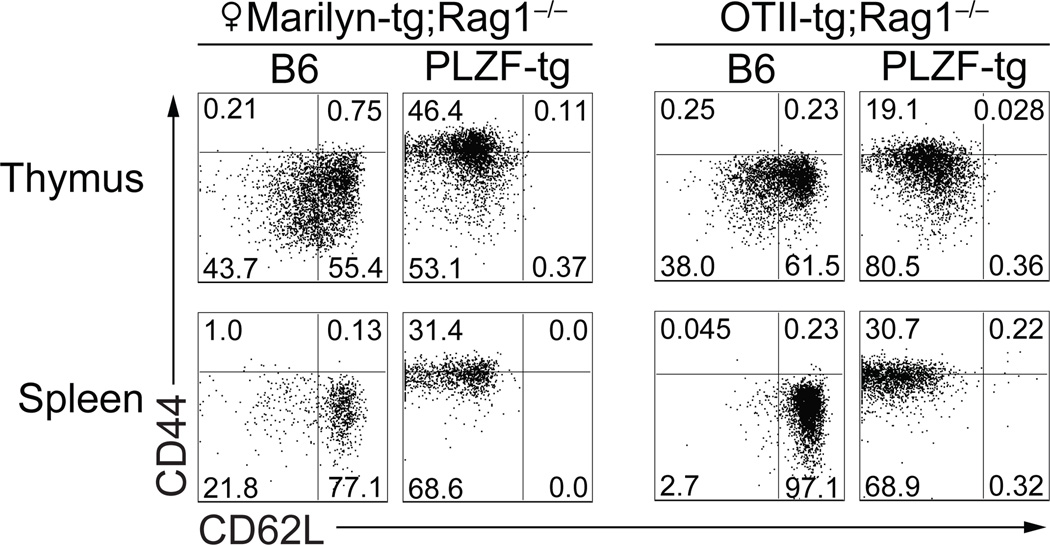
Next we introduced the PLZF transgene into the I-Ab-restricted Marilyn (HY-specific) (12) or OT-II (ovalbumin peptide-specific) (19) TCR transgenics, both in a Rag1−/− background to avoid confounding effects associated with expression of endogenous TCRs. In both cases, the FACS dot plots showed conversion of the CD4 population to a CD44hiCD62Llo phenotype (Fig. 3). Judging by CD62L expression, which was abrogated in >99.5% of CD4 cells, effector conversion involved essentially all CD4 cells in the thymus and in the periphery. As Marilyn and OT-II are well-established models for the development of conventional CD4 cells, the striking changes induced by PLZF expression demonstrated that PLZF could exert its effects downstream of conventional, non-agonist TCR/MHC II ligand interactions.
Figure 3. PLZF expression converts conventional TCR-transgenic CD4 cells into effectors.
Marilyn (I-Ab-HY specific) and OTII (I-Ab-OVA specific) TCR-transgenic mice in a Rag1−/− background were analyzed at 4–8 weeks for expression of CD44 and CD62L on CD4+Vβ6+ (Marilyn) or CD4+Vβ5+ (OTII) thymocytes and splenocytes. 3/3 Marilyn and 4/4 OTII mice examined showed similar effector conversion.
Extensive TCR repertoire overlap in wild-type and PLZF-transgenic thymocytes
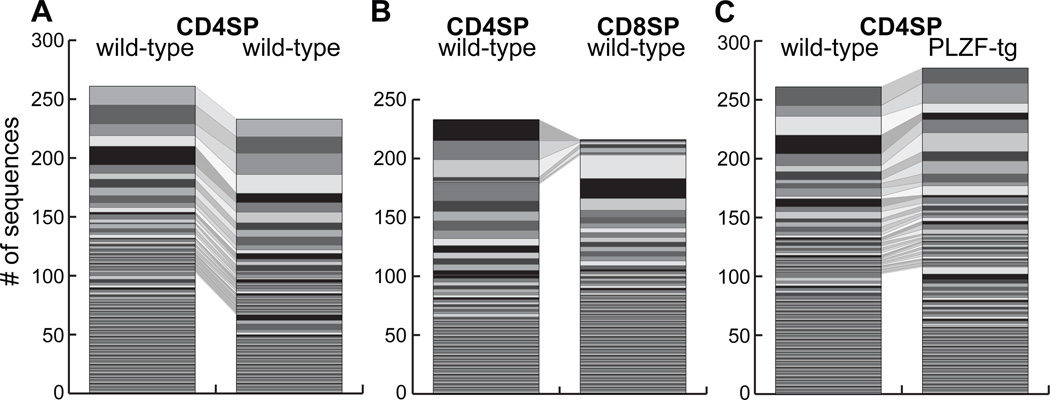
Because PLZF-transgenic effector-type cells compete equally with wild-type cells in mixed bone marrow chimeras without undergoing obvious clonal expansion (7), the conversion process must involve a large population of TCRs. This is consistent with the diversity of their Vβ usage (11). To compare the fine composition of the TCR repertoire emerging from positive and negative selection in wild type vs PLZF-transgenic cells, we fixed one TCR chain by using a Vβ7 transgene and sequenced the Vα2 chains on a large scale. This method exponentially reduces the diversity of the TCRs expressed by T cells, to the point where a large fraction of Vα2 sequences overlap between individual mice (16). As expected, we found minimal overlap between CD4 and CD8 cells (6.5%), and we confirmed the large overlap of unique sequences between CD4 thymocytes obtained from B6 littermates (38.6% on average) (Fig. 4A–C). Strikingly, there was a similarly large overlap of unique sequences between CD4 cells in WT and PLZF-transgenic littermates (33.3% overlap on average) (Fig. 4C). Table I lists the 38 sequences that were found at least 3 times in any of the CD4 data sets obtained from 2 WT and 2 PLZF-Tg (each including >200 sequences). The table demonstrates the extensive overlap between these different samples, irrespective of PLZF expression, since 31/38 sequences were found in both WT and PLZF-Tg. These findings, together with the TCR transgenic experiments, provide compelling evidence that PLZF-Tg cells expressed fundamentally the same TCR repertoire as WT, arguing against a substantial influence of PLZF in positive selection. Rather, they suggest that PLZF could act downstream of TCR-mediated selection events to impart effector differentiation to the same T cells that would have otherwise emerged as naive cells.
Figure 4. PLZF expression does not alter the TCR repertoire.
Vβ7-transgenic CD4+CD8− (CD4SP) or CD4−CD8+ (CD8SP) thymocytes on a WT or PLZF-transgenic background were FACS sorted to isolate Vα2 expressing cells and determine individual Vα2-Cα junctional sequences. Between 216 and 277 sequences were determined for each population and stacked in a bar graph where horizontal slices represent different aminoacid sequences with the height corresponding to the number of instances of that sequence; identical sequences found in different populations are connected and in the same color. (D) CD4SP thymocytes of two WT mice (average 38.6% shared unique sequences); (E), CD4SP and CD8SP thymocytes of a single WT mouse (average 6.5% shared unique sequences); (F), CD4SP thymocytes of a PLZF-transgenic and a WT littermate (average 33.3% shared unique sequences). Averages are calculated from two separate experiments including a pair of WT and transgenic littermates.
Table 1. Distribution of TCRα chains found at high frequencies among CD4 SP thymocytes.
“WT” are Vβ7-tg and “PLZF-transgenic” are Vβ7-tg PLZF-tg. Sequences found at 3 or more copies are shown (from a total of 226–277 sequences/sample).
| wild-type | PLZF-transgenic | ||||
|---|---|---|---|---|---|
| Amino acid sequence | Jα chain | exp1 | exp1 | exp1 | exp2 |
| CAARNQGGSAKLIF | TRAJ57*01 | 9 | 16 | 17 | 14 |
| CAASLNYNQGKLIF | TRAJ23*01 | 16 | 14 | 13 | 3 |
| CAASMNYNQGKLIF | TRAJ23*01 | 10 | 18 | 11 | 5 |
| CAARTGGNNKLTF | TRAJ56*01 | 5 | 9 | 16 | 14 |
| CAASANYNQGKLIF | TRAJ23*01 | 16 | 15 | 6 | 6 |
| CAASVNYNQGKLIF | TRAJ23*01 | 16 | 8 | 8 | 2 |
| CAARGSNAKLTF | TRAJ42*01 | 7 | 6 | 8 | 2 |
| CAASADYNQGKLIF | TRAJ23*01 | 7 | 8 | 3 | 3 |
| CAADNNAGAKLTF | TRAJ39*01 | 2 | 5 | 8 | 6 |
| CAASRDYNQGKLIF | TRAJ23*01 | 6 | 7 | 2 | 4 |
| CAASPNYNQGKLIF | TRAJ23*01 | 4 | 3 | 7 | 5 |
| CAASEDYNQGKLIF | TRAJ23*01 | 7 | 6 | 2 | 2 |
| CAAKNQGGSAKLIF | TRAJ57*01 | 3 | 1 | 11 | 2 |
| CAASENYNQGKLIF | TRAJ23*01 | 4 | 4 | 5 | 2 |
| CAARNNAGAKLTF | TRAJ39*01 | 0 | 5 | 6 | 3 |
| CAASEGYNQGKLIF | TRAJ23*01 | 1 | 5 | 2 | 4 |
| CAARAGGSNAKLTF | TRAJ42*01 | 3 | 2 | 4 | 2 |
| CAASARGSNAKLTF | TRAJ42*01 | 0 | 2 | 5 | 2 |
| CAASGDTGNYKYVF | TRAJ40*01 | 4 | 2 | 2 | 0 |
| CAASRGGRALIF | TRAJ15*01 | 2 | 3 | 1 | 2 |
| CAARNSNNRIFF | TRAJ31*01 | 4 | 1 | 2 | 0 |
| CAASGNYNQGKLIF | TRAJ23*01 | 3 | 3 | 0 | 1 |
| CAASYYAQGLTF | TRAJ26*01 | 2 | 1 | 1 | 3 |
| CAAKDQGGSAKLIF | TRAJ57*01 | 2 | 0 | 3 | 1 |
| CAAKGNAGAKLTF | TRAJ39*01 | 1 | 0 | 4 | 1 |
| CAARAGGNNKLTF | TRAJ56*01 | 1 | 1 | 4 | 0 |
| CAASLDYNQGKLIF | TRAJ23*01 | 3 | 0 | 2 | 0 |
| CAARYTGNYKYVF | TRAJ40*01 | 2 | 0 | 3 | 0 |
| CAASYSNNRIFF | TRAJ31*01 | 0 | 2 | 3 | 0 |
| CAAKGGSAKLIF | TRAJ57*01 | 1 | 3 | 0 | 1 |
| CAASASSGSWQLIF | TRAJ22*01 | 0 | 0 | 2 | 3 |
| CAASENTGKLTF | TRAJ27*01 | 3 | 0 | 1 | 0 |
| CAASGDTNTGKLTF | TRAJ27*01 | 3 | 0 | 0 | 0 |
| CAARRINSGGSNAKLTF | TRAJ42*01 | 0 | 0 | 3 | 0 |
| CAASWDNNAGAKLTF | TRAJ39*01 | 0 | 0 | 3 | 0 |
| CAARGGSAKLIF | TRAJ57*01 | 0 | 3 | 0 | 0 |
| CAASTNYNQGKLIF | TRAJ23*01 | 0 | 3 | 0 | 0 |
| CAAASSGSWQLIF | TRAJ22*01 | 0 | 0 | 0 | 3 |
Impact of PLZF expression on negative selection
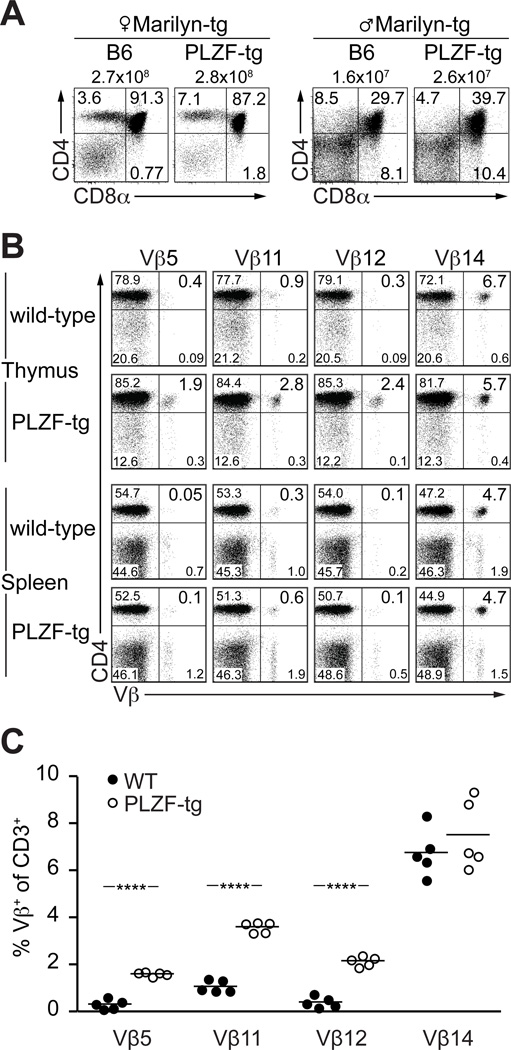
While positive selection is a major force shaping the TCR repertoire, we also investigated whether PLZF might alter negative selection of autoreactive thymocytes. I-Ab-HY-specific Marilyn TCR transgenic thymocytes undergo deletion at the DP stage of development in male mice. This process seemed unaltered by transgenic expression of PLZF, as shown by the massive reduction in thymic cellularity from >2×108 cells to ~2×107 cells in both WT and PLZF-transgenic Marilyn male mice (Fig. 5A). Because negative selection in HY TCR transgenic males may occur prior to expression of the PLZF transgene, we studied negative selection by retroviral superantigens, which occurs later during the transition from DP to SP thymocytes. We analyzed the impact of multiple endogenous retroviral superantigens by crossing WT or PLZF-transgenic B6 mice to BALB/c. In the F1 offspring, the Vβ families associated with recognition of BALB/c superantigens, including Vβ5, Vβ11 and Vβ12 were negatively selected, reduced to <1% in WT, whereas the frequency of control Vβ14 was 7% (Fig. 5B–C). In contrast, PLZF-transgenic CD4 thymocytes showed a highly significant residual frequency of 2–4% autoreactive cells expressing Vβ5, Vβ11 or Vβ12, suggesting a defect in negative selection. Importantly, however, autoreactive cells were further deleted in the periphery, suggesting that PLZF delayed rather than abrogated negative selection (Fig. 5B, compare thymus and spleen). Collectively, these findings indicated that PLZF could partially interfere with the elimination of some autoreactive thymocytes.
Figure 5. Negative selection in PLZF-transgenic mice.
(A) Influence of PLZF expression in the thymus of Marilyn (I-Ab-HY specific) TCR transgenic male and female mice examined at 4.5 weeks of age. Numbers above the dot plots indicate thymic cellularity. Data is representative of two experiments. (B) Influence of PLZF on endogenous retroviral superantigen-induced deletion of Vβ-expressing thymocytes and splenocytes. 4–8 week-old C57BL/6 × BALB/c F1 hybrid mice were analyzed for the expression of Vβ families among CD3hi thymocytes and splenocytes. F1 mice delete Vβ5, Vβ11 and Vβ12, but not Vβ14, due to endogenous viral superantigens in the BALB/c background. (C) Summary plots showing the percentage of Vβ-expressing CD3+CD4+CD8− thymocytes in individual mice as depicted in B. ****p<0.0001.
PLZF does not alter TCR-induced CD69 induction by DP thymocytes
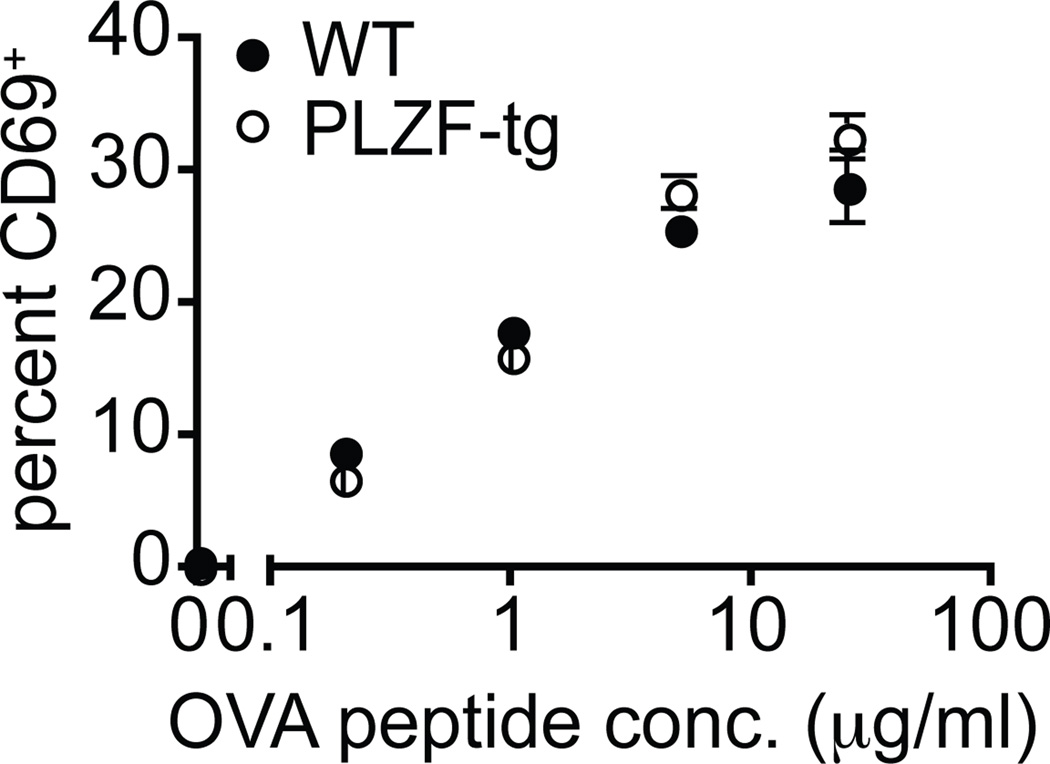
A possible explanation to the altered negative selection of PLZF transgenic thymocytes would be that PLZF somehow reduced the TCR signaling induced by agonist recognition, thus increasing the antigen concentration threshold for negative selection. We asked whether PLZF expression impacted TCR signaling by stimulating OTII DP thymocytes with graded doses of agonist OVA peptides in the presence of splenic antigen presenting cells. Prior to stimulation, the thymocytes were MACS-depleted of cells expressing CD69 to remove cells that had already been signaled in vivo (20). After stimulation with agonist peptide, CD69 was induced at the same level in both WT and PLZF-transgenic DP thymocytes over the entire range of peptide concentrations (Fig. 6). This result suggests a lack of influence of PLZF on TCR signaling during thymic selection events at the DP stage and may be consistent with a report that PLZF could enhance cell survival (21).
Figure 6. PLZF does not alter TCR signaling in DP thymocytes.
CD69-depleted thymocytes from either PLZF-transgenic OTII-transgenic Rag1−/− or OTII-transgenic Rag1+/− were stimulated with T-cell depleted splenocytes and cultured for 20 hours with varying concentrations of OTII peptide (0, 0.2, 1, 5, 25 mg/ml). The chart shows the mean percentage of CD69+ cells (from duplicate wells) among CD4+CD8+ cells. The standard error of the mean is indicated by the error bars. Data are representative of two independent experiments conducted using littermates at 4 and 8 weeks of age.
Discussion
Agonist TCR signaling, cell division and effector differentiation are generally viewed as intimately associated processes that follow recognition of foreign antigen for peripheral T cell activation (22) or of endogenous CD1d ligands for the development of autoreactive NKT cells (3). However, studies of the NKT cell-specific transcription factor PLZF demonstrate that acquisition of the effector program can be formally dissociated not only from cell division (7, 11), as previously suggested in other systems (23, 24), but also, surprisingly, from agonist signaling.
Indeed, transgenic expression of PLZF under the CD4 promoter led to the developmental induction of an effector program in CD4 SP thymocytes and thymic emigrants with all the hallmarks of conventional MHC class II-restricted T cells. They depended on MHC class II expression by radio-resistant host cells and showed extensive TCR α sequence overlap with conventional naÔve CD4 T cells when the TCR β chain was fixed. Furthermore, PLZF induced effector conversion in 2/2 MHC-class II-restricted RAG−/− TCR transgenic models examined. Collectively, these results imply that the pools of MHC class II/peptide ligands and αβ TCRs involved in the selection of wild-type and PLZF-transgenic thymocytes were largely identical and therefore that effector conversion occurred in the absence of agonist signaling.
Interestingly, it is negative selection that was impaired in the presence of PLZF. CD4 SP thymocytes expressing several retroviral superantigen-reactive Vβs exhibited a partial, but highly significant defect in negative selection, which was corrected after emigration to the periphery. These defects were not reported in a recent study of PLZF-transgenic mice because the more informative BALB/c background was not examined (11). In contrast to retroviral superantigens, the male antigen HY appeared to normally induce negative selection of HY-specific TCR transgenic thymocytes even in PLZF-transgenic animals. This may be related to different strength of TCR signaling. Alternatively, since PLZF was expressed under the CD4 promoter, the discordance could be due to different timing of deletion at the DN/early DP stage for HY TCR transgenic cells (12, 25) vs the DP to SP transition for superantigen-reactive thymocytes (26).
Importantly, PLZF did not appear to alter the TCR signaling threshold of DP thymocytes as judged by CD69 induction upon graded TCR stimulation by a range of antigen concentrations. It is possible that other signaling defects might be involved at the DP stage or at later stages during the peripheral deletion of superantigen-reactive cells. Alternatively, PLZF might exert its effect on negative selection through some anti-apoptotic function as shown in other model systems (21).
The general implication of this study is that PLZF can act as a direct switch to turn on the effector program in thymocytes that would otherwise have been selected as naÔve cells. In the context of the development of autoreactive NKT cells, where progenitors have been suggested to recognize agonist rather than partial agonist ligands, we envision a scenario whereby TCR agonist signaling and costimulation by Slamf/SAP govern the clonal expansion of NKT cells (27), whereas PLZF directly induces the acquisition of the effector program independently of agonist signaling or cell division. In addition, PLZF may favor the survival of agonist-stimulated thymocytes, suggesting some synergy between these otherwise separate pathways.
PLZF represents the first example of a transcription factor that can function as a switch to turn on the effector program without a requirement for agonist TCR signaling and cell division. Whether similar transcription factors operate during the acquisition of various effector/memory T cell programs is an intriguing possibility that warrants further investigation.
Acknowledgements
We thank members of the Bendelac laboratory for discussions, advice and help at various stages of the study; Polly Matzinger for gifts of mice and reagents; Ryan Duggan, David Leclerc, Mike Olson, and James Cao for expert assistance with cell sorting; the University of Chicago DNA Sequencing Facility, and Animal Resource Center.
This work was supported by NIH grant AI038339. A.K.S. is the recipient of the Harper Fellowship of the University of Chicago and A.B. is a Howard Hughes Medical Institute Investigator.
Abbreviations used in this paper
- PLZF
promyelocytic leukemia zinc finger
- BTB-POZ
broad complex, tramtrack, bric-a-brac, poxvirus and zinc finger
References
- 1.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 2.Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 4.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 5.Neill DR, Wong SH, Bellosi A, Flynn RJ, Daly M, Langford TK, Bucks C, Kane CM, Fallon PG, Pannell R, Jolin HE, McKenzie AN. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 2010;464:1367–1370. doi: 10.1038/nature08900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saenz SA, Siracusa MC, Perrigoue JG, Spencer SP, Urban JF, Jr, Tocker JE, Budelsky AL, Kleinschek MA, Kastelein RA, Kambayashi T, Bhandoola A, Artis D. IL25 elicits a multipotent progenitor cell population that promotes T(H)2 cytokine responses. Nature. 2010;464:1362–1366. doi: 10.1038/nature08901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li B, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. Proc Natl Acad Sci U S A. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant'Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 12.Lantz O, Grandjean I, Matzinger P, Di Santo JP. Gamma chain required for naive CD4+ T cell survival but not for antigen proliferation. Nat Immunol. 2000;1:54–58. doi: 10.1038/76917. [DOI] [PubMed] [Google Scholar]
- 13.Yu W, Nagaoka H, Jankovic M, Misulovin Z, Suh H, Rolink A, Melchers F, Meffre E, Nussenzweig MC. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization. Nature. 1999;400:682–687. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 14.Lantz O, Bendelac A. An invariant T cell receptor α chain is used by a unique subset of MHC class I-specific CD4+ and CD4−8− T cells in mice and humans. J. Exp. Med. 1994;180:1097–1106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kouskoff V, Signorelli K, Benoist C, Mathis D. Cassette vectors directing expression of T cell receptor genes in transgenic mice. J Immunol Methods. 1995;180:273–280. doi: 10.1016/0022-1759(95)00002-r. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 18.Boursalian TE, Golob J, Soper DM, Cooper CJ, Fink PJ. Continued maturation of thymic emigrants in the periphery. Nat Immunol. 2004;5:418–425. doi: 10.1038/ni1049. [DOI] [PubMed] [Google Scholar]
- 19.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76:34–40. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 20.Bendelac A, Matzinger P, Seder RA, Paul WE, Schwartz RH. Activation events during thymic selection. J. Exp. Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasim M, Carlet M, Mansha M, Greil R, Ploner C, Trockenbacher A, Rainer J, Kofler R. PLZF/ZBTB16, a glucocorticoid response gene in acute lymphoblastic leukemia, interferes with glucocorticoid-induced apoptosis. J Steroid Biochem Mol Biol. 2010;120:218–227. doi: 10.1016/j.jsbmb.2010.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Sasson SZ, Gerstel R, Hu-Li J, Paul WE. Cell division is not a "clock" measuring acquisition of competence to produce IFN-gamma or IL-4. J Immunol. 2001;166:112–120. doi: 10.4049/jimmunol.166.1.112. [DOI] [PubMed] [Google Scholar]
- 24.Laouar Y, Crispe IN. Functional flexibility in T cells: independent regulation of CD4+ T cell proliferation and effector function in vivo. Immunity. 2000;13:291–301. doi: 10.1016/s1074-7613(00)00029-7. [DOI] [PubMed] [Google Scholar]
- 25.Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- 26.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 27.Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]