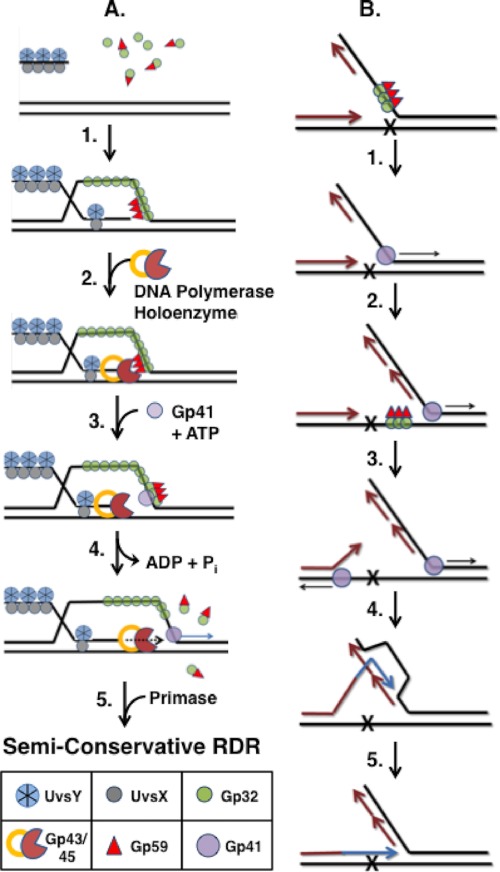
FIGURE 8.
Models for Gp59-dependent helicase loading at Gp32-ssDNA clusters during recombination-dependent DNA replication and repair. A, strand-specific loading of Gp41 helicase during RDR (Step 1). Following DNA strand invasion promoted by UvsX recombinase, UvsY mediator, and Gp32, Gp59 co-localizes with Gp32 clusters on the displaced strand of the D-loop intermediate, forming a helicase loading complex. The structure-specific DNA binding activity of Gp59 may help target the HLC to the leading edge (fork junction) of the D-loop (Step 2). As the protein components of the T4 replisome assemble, the presence of Gp59 at the fork inhibits Gp43 (DNA polymerase) from replicating the leading strand (Step 3). Recruitment of Gp41-ATP rearranges the HLC so that it no longer blocks the leading strand polymerase (Step 4). ATP hydrolysis by Gp41 drives unwinding of the duplex ahead of Gp43. Gp32 and Gp59 both are ejected from the helicase assembly site but may be recycled for additional rounds of helicase loading (see text for details) (Step 5). Primase recruitment reconstitutes the Gp41/Gp61 primosome leading to semiconservative RDR. B, hypothetical model for multiple rounds of helicase loading during error-free lesion bypass. This process involves the uncoupling of leading/lagging strand DNA synthesis followed by a round of RDR using a newly synthesized Okazaki fragment as template. Newly synthesized DNA is shown as either magenta (semiconservative synthesis) or blue (bubble migration synthesis). Replication and recombination proteins other than Gp59, Gp32, and Gp41 are omitted for clarity (Step 1). Leading and lagging strand syntheses are initially coupled until the leading strand polymerase encounters a lesion and becomes stalled. Uncoupling of leading/lagging strand synthesis could require a new round of helicase loading by Gp59 targeting Gp32 clusters on the lagging strand ssDNA (Step 2). Continued helicase translocation allows for the synthesis of a new Okazaki fragment overlapping the site of damage. Helicase activity also exposes ssDNA on the damaged strand that is sufficiently long for HLC assembly (Step 3). An additional round of helicase loading and translocation from this site leads to the unwinding of the stalled daughter strand from the damaged template (Step 4). The displaced 3′-end of the daughter strand primes a round of RDR using the overlapping Okazaki fragment as a template (Step 5). Bubble migration RDR extends the daughter strand until it can reanneal to the original template and continue semiconservative synthesis, having bypassed the lesion. See text for details.