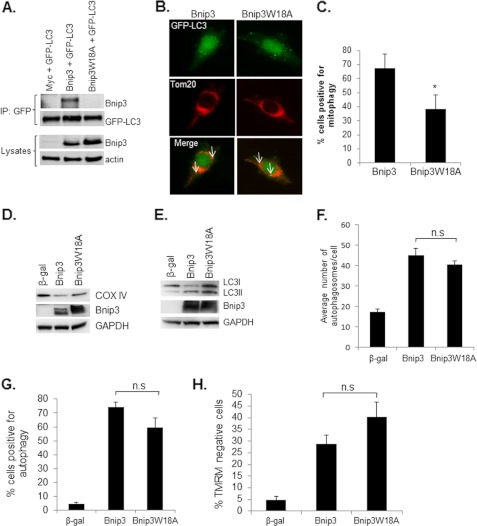
FIGURE 3.
Disrupting the interaction between Bnip3 and LC3 reduces mitophagy. A, a point mutation in the conserved LIR disrupts the interaction between Bnip3 and LC3. Bnip3W18A failed to coimmunoprecipitate (IP) with LC3 in HeLa cells. Equal expression levels of Bnip3 and Bnip3W18A were confirmed by Western blot analysis. Actin was used as a loading control. B, representative images of HeLa cells infected with Bnip3 or Bnip3W18A plus GFP-LC3. Mitophagy was determined by assessing colocalization (white arrow) between GFP-LC3-positive autophagosomes and Tom20-labeled mitochondria using fluorescence microscopy. C, quantitation of the number of cells positive for mitophagy. Bnip3W18A induced significantly less mitophagy compared with Bnip3. *, p < 0.05. n = 4. D, mitophagy was also confirmed by Western blotting for cytochrome c oxidase subunit IV (COX IV), a mitochondrial inner membrane protein, in HeLa cells infected with β-gal, Bnip3, or Bnip3W18A. GAPDH was used as a loading control. E, Western blot analysis for endogenous LC3I/II. F, quantitation of the number of GFP-LC3-positive autophagosomes per cell. n = 4. n.s., not significant. G, HeLa cells overexpressing β-gal, Bnip3, or Bnip3W18A were scored for induction of autophagy (n = 3). H, HeLa cells overexpressing β-gal, Bnip3, or Bnip3W18A stained with tetramethylrhodamine methyl ester (TMRM) to measure mitochondrial membrane potential (n = 4).