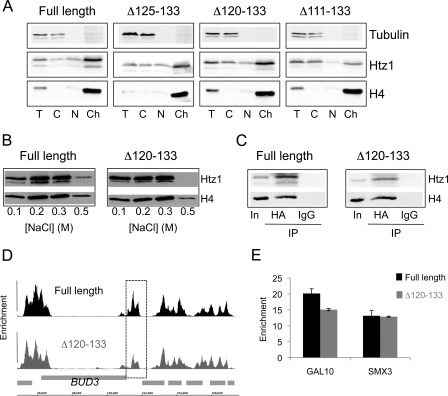
FIGURE 3.
The C terminus is required for stable retention of Htz1 in chromatin but not for normal deposition. A, proteins from various sub-cellular fractions (T, total; C, cytoplasm; n, nuclear; Ch, chromatin) derived from the indicated strains were analyzed by Western blotting. Membranes were probed with anti-HA antibodies to detect Htz1 and its truncated forms; anti-H4 and anti-tubulin were included to verify expected fractionation of proteins. B, chromatin fraction from wild-type (full-length) or Htz1Δ120–133 cells was washed with buffers containing various salt concentrations (indicated below), and the proteins remaining in the pellet were assayed by Western blotting. C, immunoprecipitation of HA-Htz1 proteins from the soluble, primarily mononucleosomal, fraction produced by MNase digestion (input; In) results in co-precipitation of H4. Immunoprecipitation with an unrelated antibody (IgG) does not precipitate H4. D, view of a region of chromosome III (coordinates indicated at bottom) displaying the enrichments of HA-Htz1 (top; black) and HA-htz1Δ120–133 (bottom; dark gray) relative to S. cerevisiae open reading frames (light gray boxes). This region around the BUD3 gene is representative of the pattern across the entire genome and was chosen for ease of comparison to previously published data (37). The dashed box surrounds a region with lower Htz1 occupancy in the Δ120–133 mutant. E, Q-PCR analysis of ChIP enrichments of full-length Htz1 and Htz1Δ120–133 at the GAL10 and SMX3 genes relative to a non-enriched control region at HMR.