Abstract
Atrial fibrillation is a highly prevalent arrhythmia and a major risk factor for stroke, heart failure and death1. We conducted a genome-wide association study (GWAS) in individuals of European ancestry, including 6,707 with and 52,426 without atrial fibrillation. Six new atrial fibrillation susceptibility loci were identified and replicated in an additional sample of individuals of European ancestry, including 5,381 subjects with and 1 0,030 subjects without atrial fibrillation (P < 5 × 10−8). Four of the loci identified in Europeans were further replicated in silico in a GWAS of Japanese individuals, including 843 individuals with and 3,350 individuals without atrial fibrillation. The identified loci implicate candidate genes that encode transcription factors related to cardiopulmonary development, cardiac-expressed ion channels and cell signaling molecules.
Genome-wide association studies in individuals of European descent have identified three genomic regions associated with atrial fibrillation on chromosomes 4q25 (PITX2)2–5, 16q22 (ZFHX3)3,5 and 1q21 (KCNN3)2. Studies of electrocardiographic traits have also identified a number of loci associated with atrial fibrillation6,7. However, despite these findings, much of the heritability of atrial fibrillation remains unexplained, justifying the search for additional genetic variants underlying atrial fibrillation risk8. Large-scale meta-analysis of GWAS results is a powerful method to identify additional genetic variation underlying traits and conditions. We therefore conducted a meta-analysis of multiple well-phenotyped GWAS samples of European ancestry to identify additional atrial fibrillation susceptibility loci.
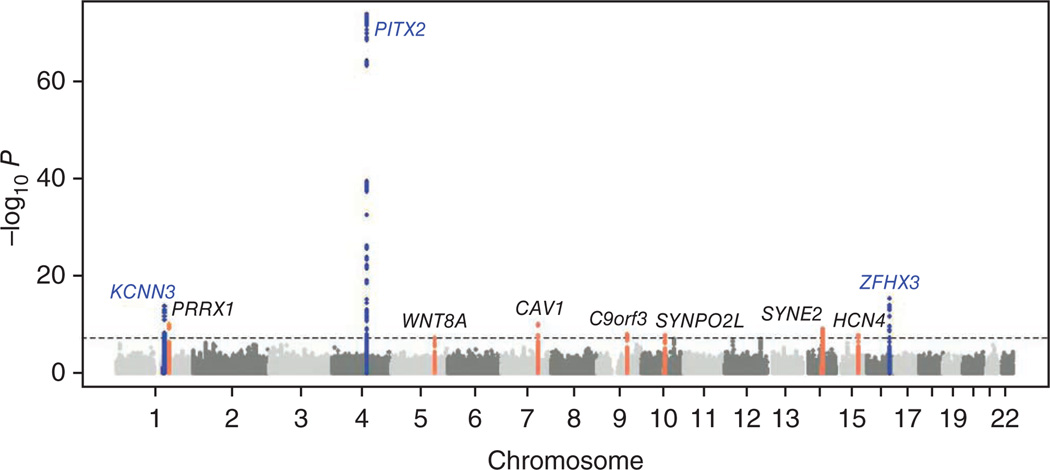
Six prospective cohort and 10 prevalent study samples contributed to the discovery analysis, which was adjusted for age and sex (Table 1, Online Methods and Supplementary Note). Atrial fibrillation status was systematically ascertained in each sample (Online Methods and Supplementary Note). After application of quality control SNP exclusion criteria in each study (Supplementary Table 1), meta-analysis was performed, applying genomic control to each study. The genomic control inflation factor for the meta-analysis was 1.042 for the full set of SNPs and 1.040 after omitting all SNPs within 500 kb of the association signals that reached genome-wide significance. The quantile-quantile plot of the expected versus observed P-value distributions for association of the 2,609,549 SNPs analyzed is shown (Supplementary Fig. 1). We identified ten loci that exceeded our preset threshold for genome-wide significance (P < 5 × 10−8) (Fig. 1). The three loci most significantly associated with atrial fibrillation were at previously identified atrial fibrillation susceptibility loci on chromosomes 4q25 in PITX2 (rs6817105; P = 1.8 × 10−74)4, 16q22 in ZFHX3 (rs2106261; P = 3.2 × 10−16)3,5 and 1q21 in KCNN3 (rs6666258; P = 2.0 × 10−14)2 (Table 2).
Table 1.
Subject characteristics
| Cohort | Cohort type |
Participants (n) |
AF (n) | Males (%) | Age at DNA collection (mean ± s.d.) |
Age at DNA collection (range) |
Age of AF onset (mean ± s.d.) |
Hypertension n (%) |
Body mass index, in kg/m2 (mean ± s.d.) |
Diabetes n (%) |
Myocardial infarction n (%) |
Heart failure n (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incident AF | Overall N | |||||||||||
| ARIC | Cohort | 8,890 | 802 | 4,181 (47.0) | 54.3 ± 5.7 | 44–66 | 67.8 ± 6.8 | 2,376 (26.7) | 27.0 ± 4.8 | 763 (8.6) | 354 (4.0) | 325 (3.7) |
| AGES | Cohort | 2,959 | 158 | 1,154 (39.0) | 76.5 ± 5.5 | 66–95 | 75.0 ± 8.8 | 2,595 (87.7) | 27.1 ± 4.4 | 319 (10.8) | 189 (6.39) | 55 (1.9) |
| CHS | Cohort | 3,204 | 764 | 1,242 (38.8) | 72.2 ± 5.3 | 65–98 | 81.2 ± 6.0 | 1,678 (52.4) | 26.3 ± 4.4 | 377 (11.8) | 0 | 0 |
| FHS | Cohort | 4,062 | 310 | 1,771 (43.6) | 64.7 ± 12.6 | 31–101 | 77.8 ± 10.6 | 2,001 (49.3) | 27.7 ± 5.2 | 328 (8.07) | 231 (5.7) | 55 (1.4) |
| RS-I | Cohort | 5,665 | 542 | 2,282 (40.3) | 69.1 ± 9.0 | 55–99 | 77.6 ± 7.7 | 1,866 (32.9) | 26.3 ± 3.7 | 567 (10.0) | 632 (11.2) | 156 (2.8) |
| WGHS | Cohort | 20,836 | 648 | 0 | 54.1 ± 7.0 | 43–89 | 68.0 ± 8.2 | 5,022 (24.1) | 25.9 ± 4.9 | 503 (2.4) | 0 | 17 (0.1) |
| Prevalent AF | N per group | |||||||||||
| AFNET | Cases | 468 | – | 236 (50.4) | 51.8 ± 7.2 | 29–74 | 51.3 ± 7.6 | 252 (53.8) | 28.0 ± 4.9 | 36 (7.7) | 6 (1.8) | 14 (4.8) |
| KORA | Referents | 438 | – | 219 (50.0) | 56.2 ± 7.1 | 45–69 | – | 185 (42.2) | 27.7 ± 4.5 | 37 (8.4) | 6 (1.3) | 13 (2.9) |
| AGESa | Cases | 241 | – | 88 (55.7) | 78.5 ± 5.9 | 67–95 | 80.9 ± 6.2 | 143 (90.5) | 27.7 ± 4.4 | 20 (12.7) | 8 (5.1) | 5 (3.2) |
| Referents | 2,718 | – | 70 (36.1) | 76.1 ± 5.4 | 66–94 | 80.4 ± 5.4 | 2,002 (78.2) | 27.0 ± 4.5 | 269 (10.5) | 122 (4.8) | 27 (1.1) | |
| CC | Cases | 496 | – | 375 (75.6) | 58.8 ± 10.7 | 20–84 | 51.7 ± 12.0 | 269 (54.2) | 30.2 ± 6.2 | 28 (5.6) | 0 (0) | 0 (0) |
| Referents | 2,971 | – | 1,124 (37.8) | 28.5 ± 22.2 | 0–87 | – | – | – | – | – | – | |
| HVH | Cases | 95 | – | 28 (29.5) | 59.5 ± 6.5 | 40–68 | 57.4 ± 6.4 | 50 (52.6) | 34.1 ± 9.9 | 14 (14.7) | 0 | 0 |
| Referents | 193 | – | 106 (54.9) | 59.5 ± 6.0 | 40–69 | – | 153 (79.3) | 31.4 ± 7.2 | 31 (16.1) | 9 (4.7) | 7 (3.6) | |
| CHSa | Cases | 67 | – | 38 (56.7) | 76.3 ± 5.8 | 66–90 | – | 35 (52.2) | 26.6 ± 4.3 | 14 (20.9) | 0 | 0 |
| Referents | 3,204 | – | 1,242 (38.8) | 72.2 ± 5.3 | 65–98 | – | 1,678 (52.4) | 26.3 ± 4.4 | 377 (11.8) | 0 | 0 | |
| FHSa | Cases | 253 | – | 151 (59.7) | 76.9 ± 9.9 | 45–97 | 70.9 ± 10.8 | 180 (71.1) | 27.4 ± 4.8 | 41 (16.21) | 60 (23.7) | 57 (0.23) |
| Referents | 4,151 | – | 1,807 (43.5) | 64.7 ± 12.6 | 31–101 | – | 2,036 (49.1) | 27.7 ± 5.2 | 329 (7.9) | 235 (5.7) | 55 (1.32) | |
| MGH | Cases | 366 | – | 295 (80.6) | 53.4 ± 10.5 | 21–77 | 46.1 ± 11.7 | 85.8 (22.7) | 27.8 ± 5.0 | 12 (3.2) | 4 (1.1) | 10 (2.8) |
| MIGEN | Referents | 911 | – | 485 (53.2) | 47.9 ± 8.8 | 18–83 | – | – | – | – | – | – |
| RS-Ia | Cases | 309 | – | 145 (46.9) | 76.2 ± 8.7 | 56–98 | – | 131 (42.4) | 25.9 ± 3.6 | 64 (20.7) | 69 (22.3) | 54 (17.5) |
| Referents | 5,665 | – | 2,282 (40.3) | 69.1 ± 9.0 | 55–99 | – | 1,866 (32.9) | 26.3 ± 3.7 | 567 (10.0) | 632 (11.2) | 156 (2.8) | |
| SHIP | Cases | 107 | – | 69 (64.5) | 65.1 ± 11.5 | 21–81 | – | 59 (55.1) | 29.6 ± 5.1 | 23 (21.5) | 14 (13.1) | 44 (41.1) |
| Referents | 1,816 | – | 906 (49.9) | 50.7 ± 14.9 | 21–81 | – | 437 (24.1) | 27.2 ± 4.5 | 131 (7.2) | 54 (3.0) | 157 (8.6) | |
| Vanderbilt | Cases | 1,081 | – | 738 (68.3) | 59.5 ± 12.6 | 16–87 | 51.5 ± 14.7 | 625 (57.8) | 30.7 ± 6.9 | 197 (18.2) | 95 (8.8) | 191 (17.6) |
| Referents | 880 | – | 551 (62.6) | 50.0 ± 17.4 | 18–91 | – | 463 (52.6) | 28.0 ± 5.8 | 180 (20.5) | 299 (34.0) | 120 (13.6) |
AF, atrial fibrillation.
Studies included community-based prospectively ascertained prevalent atrial fibrillation cases. The other prevalent atrial fibrillation studies were of a case-control design.
Figure 1.
Manhattan plot of meta-analysis results for genome-wide association with atrial fibrillation. The −log10 (P value) is plotted against the physical position of each SNP on each chromosome. The threshold for genome-wide significance, P < 5 × 10−8, is indicated by the dashed line. The three previously reported loci for atrial fibrillation are indicated in blue, and the seven new loci that exceeded the genome-wide significance threshold are indicated in orange.
Table 2.
Summary of GWAS meta-analysis results with P < 5 × 10−8
| Discovery | Replication | Overall | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Locus | Closest gene |
SNP location relative to closest gene |
Minor/ major allele |
MAF (%) |
RR (95% CI) |
Meta P value |
I2 (%), P value |
RR (95% CI) |
Meta P value |
RR (95% CI) |
Meta P value |
| rs6666258 | 1q21 | KCNN3-PMVK | Intronic | C/G | 29.9 | 1.18 (1.13–1.23) | 2.0 × 10−14 | 42.3, 0.04 | – | – | – | – |
| rs3903239 | 1q24 | PRRX1 | 46 kb upstream | G/A | 44.7 | 1.14 (1.10–1.18) | 9.1 × 10−11 | 53.2, 6.3 × 10−3 | 1.13 (1.06–1.20) | 2.0 × 10−4 | 1.14 (1.10–1.17) | 8.4 × 10−14 |
| rs6817105 | 4q25 | PITX2 | 150 kb upstream | C/T | 13.1 | 1.64 (1.55–1.73) | 1.8 × 10−74 | 80.8, 1.4 × 10−10 | – | – | – | – |
| rs2040862 | 5q31 | WNT8A | Intronic | T/C | 17.8 | 1.15 (1.09–1.21) | 3.2 × 10−8 | 10, 0.34 | 1.04 (0.96–1.12) | 3.6 × 10−1 | 1.12 (1.07–1.17) | 2.5 × 10−7 |
| rs3807989 | 7q31 | CAV1 | Intronic | A/G | 40.4 | 0.88 (0.84–0.91) | 9.6 × 10−11 | 10, 0.34 | 0.93 (0.88–0.97) | 2.7 × 10−3 | 0.90 (0.87–0.92) | 3.6 × 10−12 |
| rs10821415 | 9q22 | C9orf3 | Intronic | A/C | 42.4 | 1.13 (1.08–1.18) | 7.9 × 10−9 | 49.5, 0.015 | 1.09 (1.04–1.15) | 7.2 × 10−4 | 1.11 (1.08–1.15) | 4.2 × 10−11 |
| rs10824026 | 10q22 | SYNPO2L | 5 kb upstream | G/A | 15.8 | 0.85 (0.81–0.90) | 1.7 × 10−8 | 37.9, 0.06 | 0.91 (0.83–0.99) | 3.5 × 10−2 | 0.87 (0.83–0.91) | 4.0 × 10−9 |
| rs1152591 | 14q23 | SYNE2 | Intronic | A/G | 47.6 | 1.13 (1.09–1.18) | 6.2 × 10−10 | 25.7, 0.16 | 1.12 (1.06–1.19) | 1.9 × 10−4 | 1.13 (1.09–1.17) | 5.8 × 10−13 |
| rs7164883 | 15q24 | HCN4 | Intronic | G/A | 16.0 | 1.16 (1.10–1.22) | 1.3 × 10−8 | 0, 0.85 | 1.24 (1.16–1.32) | 1.3 × 10−10 | 1.19 (1.14–1.24) | 2.8 × 10−17 |
| rs2106261 | 16q22 | ZFHX3 | Intronic | T/C | 17.6 | 1.24 (1.17–1.30) | 3.2 × 10−16 | 58.8, 1.6 × 10−3 | – | – | – | – |
MAF, minor allele frequency; RR, relative risk. I2 represents the proportion of variability in the effect size due to between-study variability. We did not attempt replication of the previously published genetic loci associated with atrial fibrillation on chromosomes 1q21 (KCNN3)2, 4q25 (PITX2)4 and 16q22 (ZFHX3)3,5.
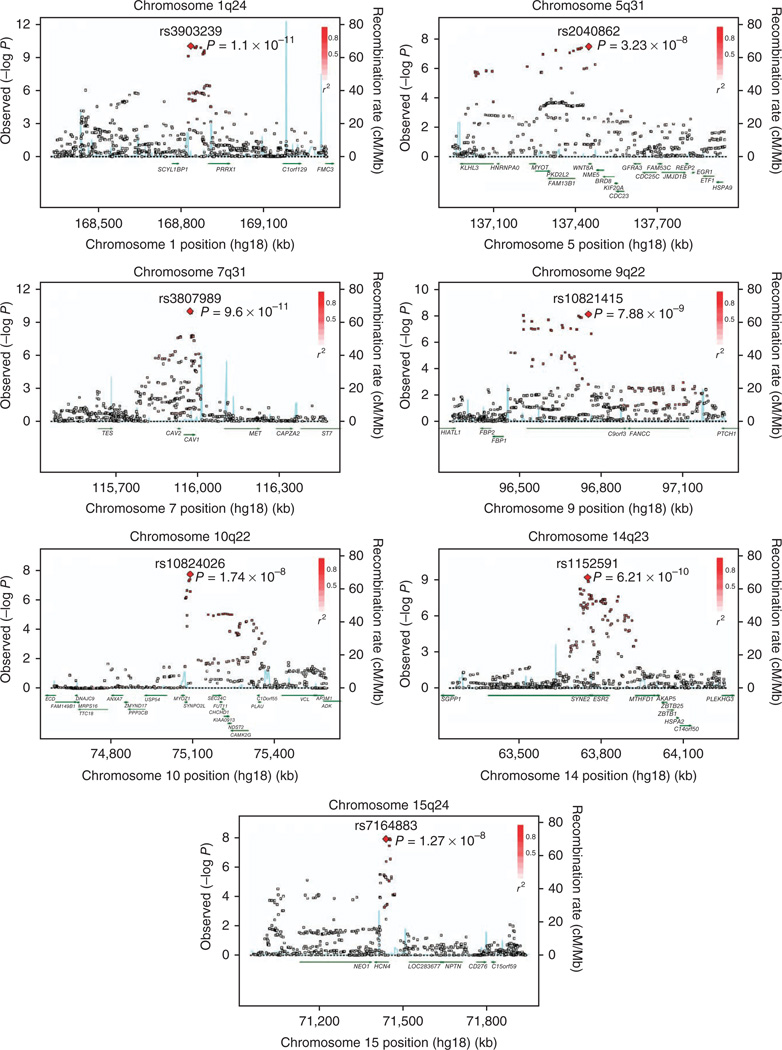
Seven new genomic loci were associated with atrial fibrillation with P < 5 × 10−8 in the discovery stage (Table 2). The most significantly associated SNP in each of the seven new loci was genotyped and tested for association with atrial fibrillation in an additional 3,132 to 5,289 independent individuals with atrial fibrillation and 8,159 to 11,148 referent individuals derived from six studies of individuals of European ancestry (Supplementary Table 2). Six of the loci associated with atrial fibrillation in the discovery stage met our criteria for independent replication. Study-specific replication results are detailed (Supplementary Table 3). The results from meta-analysis of the discovery and replication results are shown (Table 2), as are regional plots (Fig. 2). Recognizing that the genes in closest physical proximity to the associated SNPs are not always the causative genes, we report below the genetic associations in order of statistical significance along with the nearest gene.
Figure 2.
Regional plots for seven new atrial fibrillation loci in the discovery sample with P < 1 × 10−8. SNPs are plotted by meta-analysis P value and genomic position (NCBI Build 36). The SNP of interest is labeled. The strength of LD is indicated by red coloring. Estimated recombination rates are shown by the blue peaks, and gene annotations are indicated by dark green arrows. LD and recombination rates are based on the Utah residents of Northern and Western European ancestry (CEU) HapMap cohort (release 22). Plots were prepared using SNAP27.
The most significant new association in the discovery stage was on chromosome 1q24 (rs3903239; overall P = 8.4 × 10−14) in PRRX1, which encodes a homeodomain transcription factor highly expressed in the developing heart, particularly in connective tissue. Biological interaction between PRRX1 and a related homeobox transcription factor gene, PRRX2, results in abnormalities of great vessel development in a mouse knockout model9. In a separate PRRX1 knockout, fetal pulmonary vasculature development was impaired10.
A second locus was identified on chromosome 7q31 (rs3807989; overall P = 3.6 × 10−12) in CAV1, which encodes caveolin-1, a cellular membrane protein involved in signal transduction. CAV1 is selectively expressed in the atria11, and its knockout has been associated with dilated cardiomyopathy12. The CAV1 protein colocalizes with and negatively regulates the activity of KCNH2 (ref. 13), a potassium channel involved in cardiac repolarization; the corresponding KCNH2 gene was found to be associated with atrial fibrillation in a candidate gene association study, although not in our present analysis14. The top SNP at the CAV1 locus identified in the current study, rs3807989, was previously identified in a GWAS of the PR and QRS intervals and related to atrial fibrillation6,7. The relationship between other previously reported PR-associated loci and atrial fibrillation are reported (Supplementary Table 4). Of note, significant associations with atrial fibrillation were observed for SNPs related to the PR interval in SOX5, TBX5, SCN5A and SCN10A6,7.
The third locus on chromosome 14q23 (rs1152591; overall P = 5.8 × 10−13) was located in an intron of SYNE2, which encodes numerous nesprin-2 isoforms, some of which are highly expressed in the heart and skeletal muscle. Nesprin-2 localizes throughout the sarcomere and is involved in maintaining nuclear structural integrity by anchoring the nucleus to the cytoskeleton. In a candidate gene approach, mutations in SYNE2 were found to segregate in some families with Emery-Dreifuss muscular dystrophy15, which is characterized by skeletal muscle atrophy, cardiomyopathy and cardiac conduction defects.
The fourth locus on chromosome 9q22 (rs10821415; overall P = 4.2 × 1011) was located in an ORF on chromosome 9. Genes at this locus include FBP1 and FBP2, which are important for gluconeogenesis. Autosomal recessive FBP1 deficiency has been described, but cardiovascular features did not seem to be prominent16. Variants at 9q22 have been implicated in the regulation of height, pulmonary function, angiogenesis and attention deficit hyperactivity disorder, although rs10821415 is not in substantial linkage disequilibrium (LD) with any of these SNPs (r2 < 0.30).
A fifth signal was located at 15q24 (rs7164883; overall P = 2.8 × 10−17) in the first intron of HCN4. The HCN4 protein is the predominant cardiac hyperpolarization-activated cyclic nucleotide–gated channel and is highly expressed in the sinoatrial node. HCN4 activity underlies the funny current (If) that governs cardiac pacemaking, and mutations in HCN4 have been associated with various forms of sinus nodal dysfunction17,18.
A sixth locus on chromosome 10q22 (rs10824026; overall P = 4.0 × 10−9) was located 5 kb upstream of SYNPO2L and 20 kb upstream of MYOZ1. The proteins encoded by SYNPO2L and MYOZ1 are both expressed in skeletal and cardiac muscle, localize to the Z-disc and interact with numerous other proteins. However, the precise role of either gene in cardiovascular physiology is unknown19,20. A mouse knockout of MYOZ1 showed increased calcineurin activity and cardiac hypertrophy in response to pressure overload. However, candidate gene approaches have not supported a prominent role for MYOZ1 mutations in causing familial dilated cardiomyopathy21. Of note, the SYNPO2L locus is located within a previously reported atrial fibrillation susceptibility locus identified in a family with autosomal dominant atrial fibrillation22.
One other locus was identified in the meta-analysis of atrial fibrillation in the WNT8A gene (rs2040862; P = 3.2 × 10−8); however, this association failed to replicate in additional independent cohorts with atrial fibrillation (replication P = 0.36; combined P = 2.5 × 10−7).
There was evidence of significant heterogeneity in the discovery meta-analysis at the previously published atrial fibrillation susceptibility signals at 4q25 in PITX2 and at 16q22 in ZFHX3 (Table 2). Effect heterogeneity at the PITX2 locus has already been observed4,23,24.
We then sought to determine whether the top SNPs or their proxies at each locus were associated with alterations in gene expression in an expression quantitative trait locus (eQTL) database, the Genotype-Tissue Expression eQTL browser. The top SNP at the SYNPO2L locus, rs10824026, is in strong LD with a SNP, rs12570126 (R2 = 0.932), that was found to correlate with the expression of MYOZ1 and SYNPO2L (P = 1.5 × 10−6; data derived from lymphoblastoid cell lines in 270 individuals from the HapMap Consortium)25. Furthermore, the top SNP at the SYNPO2L locus is in LD with a nonsynonymous SNP in SYNPO2L, rs3812629 (R2 = 0.8; encoding the SYNPO2L P707L variant with respect to transcript Q9H987-1), that is predicted to be damaging by both the PolyPhen-2 and the SIFT algorithms. None of the other identified atrial fibrillation risk SNPs were associated with variations in gene expression in the Genotype-Tissue Expression eQTL browser.
We next examined the generalizability of our findings by examining our results in a separate GWAS of individuals of Japanese ancestry, including 843 subjects with and 3,350 subjects without atrial fibrillation in the Japan BioBank study (Supplementary Fig. 2a,b). Within the Japanese GWAS, only the chromosome 4q25 (PITX2) locus exceeded the preset threshold for genome-wide significance (rs2634073; odds ratio (OR) = 1.84, 95% confidence interval (CI) = 1.59–2.13; 3.7 × 10−17; Supplementary Figs. 2a and 3 and Supplementary Table 5). At the previously published locus at 16q22 (ZFHX3)3,5, rs12932445 was associated with atrial fibrillation in participants of Japanese ancestry (P = 6.8 × 10−4). The relationship between atrial fibrillation and variants at the KCNN3-PMVK locus on chromosome 1q21 failed to replicate (P = 0.17); however, a regional plot of this locus revealed a distinct signal at the rs7514452 SNP—approximately 375 kb away from rs6666258, the top SNP in the European ancestry sample—that was modestly associated with atrial fibrillation in the Japanese sample (P = 4.9 × 10−5; R2 = 0.002). At PRRX1 and CAV1, the top SNPs in the European samples were also associated with atrial fibrillation in Japanese individuals. For the loci near PRRX1 and C9orf3, alternate SNPs in the Japanese cohort were more significantly associated with atrial fibrillation than the top SNP at each locus in Europeans (Supplementary Fig. 3 and Supplementary Table 5).
Our study was subject to a number of limitations. To maximize both the power and the generalizability of our study, we included all available individuals with atrial fibrillation; thus, some individuals had comorbidities, such as systolic dysfunction and hypertension. However, none of the identified risk variants for atrial fibrillation were strongly associated with systolic dysfunction in the EchoGen Consortium26, a meta-analysis of echocardiographic data from 5 community-based cohorts consisting of over 12,000 individuals of European descent (P value < 1 × 10−5). Further, when our replication results were adjusted for hypertension status, the identified variants remained significantly associated with atrial fibrillation (Supplementary Table 3). Ultimately, the development of a comprehensive risk score incorporating clinical, biochemical and genetic marker data will be necessary to clarify the incremental benefit of our findings in clinical care. Our eQTL analyses were limited to data available within the Genotype-Tissue Expression eQTL browser; future eQTL analyses in cardiac tissue may be helpful in identifying a relationship between the SNPs associated with atrial fibrillation risk and gene expression. Finally, we acknowledge that the identified variants may not be causal but may represent causal elements in the same or different molecular pathways; future statistical, bioinformatic and biological analyses investigating potential genetic interactions are warranted. Fine mapping and deep resequencing will be necessary to uncover the genetic architecture accompanying the identified common atrial fibrillation susceptibility signals.
In summary, our GWAS meta-analysis for atrial fibrillation has identified six new susceptibility loci in or near plausible candidate genes involved in pacemaking activity, signal transduction and cardiopulmonary development. Our results show that atrial fibrillation has multiple genetic associations and identifies new targets for biological investigation.
URLs
BIMBAM, http://quartus.uchicago.edu/~yguan/bimbam/index.html; Genotype-Tissue Expression eQTL browser, http://www.ncbi.nlm.nih.gov/gtex/GTEX2/gtex.cgi; iControlDB database, http://www.illumina.com/science/icontroldb.ilmn; IMPUTE, https://mathgen.stats.ox.ac.uk/impute/impute.html; MACH, http://www.sph.umich.edu/csg/abecasis/MACH/; METAL, http://www.sph.umich.edu/csg/abecasis/metal/index.html; PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/; SIFT, http://sift.jcvi.org/; SNAP, http://www.broadinstitute.org/mpg/snap/ldsearch.php.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
ACKNOWLEDGMENTS
Acknowledgments are contained in the Supplementary Note.
Footnotes
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRIBUTIONS
Study concept and design was determined by P.T.E., K.L.L., C.M.A., B.P.K., M.K.C., J.D., H.V., A.H., Y.N., D.M.R., A.G.U., A.B.N., Y.L., T. Tanaka, B.H.C.S., S.B.F., D.D., S.R.H., E.J.B., V.G. and S.K. Acquisition of data was performed by P.T.E., C.M.A., N.L.G., J.C.B., M.K.C., M.D., J.D.R., A.P., M.F.S., J.D., N.L.S., J.D.S., R.W., J.I.R., L.J.L., T.B.H., U.V., A.H., E.Z.S., M.K., U.B.T., D.C., T.F., R.M., B.M.P., T.M., S.P., H.-E.W., J.C.M.W., A.G.U., F.R., M.S., A.B.N., Y.L., M.H.G., B.H.C.S., A.A., S.R.H., E.J.B., V.G. and S.K. Analysis and interpretation of data were performed by P.T.E., K.L.L., N.L.G., M.D.R., A.V.S., D.E.A., M.M.-N., B.P.K., S.A.L., J.C.B., M.D., K.O., J.D.R., J.G.S., M.F.S., K.L., J.D., N.L.S., K.M.R., J.I.R., R.D.W., U.V., G.L., T. Tsunoda, N.S., N.K., B.M.P., S.K., D.M.R., B.M., Y.L., M.H.G., D.D., S.R.H. and E.J.B. Drafting of the manuscript was carried out by P.T.E., K.L.L., B.P.K., S.A.L. and S.K. Critical revision of the manuscript for important intellectual content was carried out by C.M.A., N.L.G., A.V.S., D.E.A., B.P.K., J.C.B., M.K.C., M.D., J.G.S., A.P., M.F.S., J.D., N.L.S., J.D.S., M.R., K.M.R., D.R.V.W., J.W.M., R.W., J.I.R., L.J.L., T.B.H., U.V., H.V., D.J.M., A.H., L.Y.C., E.Z.S., G.L., U.B.T., P.M.R., D.C., N.S., S.M., K.L.F., B.M.P., T.M., S.P., J.C.M.W., D.M.R., A.G.U., F.R., B.M., M.S., A.B.N., Y.L., M.H.G., O.M., B.H.C.S., S.B.F., A.A., D.D., D.I.C., S.R.H., E.J.B. and V.G. Statistical analysis was performed by K.L.L., N.L.G., M.D.R., A.V.S., D.E.A., M.M.-N., B.P.K., J.C.B., J.G.S., M.F.S., K.L., J.D., K.M.R., R.W.D., U.V., G.L., L.M.R., Y.L. and D.I.C. Funding was obtained by P.T.E., C.M.A., M.K.C., J.G.S., N.L.S., J.D.S., J.I.R., L.J.L., T.B.H., H.V., A.H., E.B., U.B.T., P.M.R., B.M.P., T.M., H.-E.W., J.C.M.W., D.M.R., A.G.U., Y.L., O.M., B.H.C.S., S.B.F., A.A., D.D., S.R.H., E.J.B., V.G. and S.K. Study supervision was performed by P.T.E., K.L.L., C.M.A., M.K.C., N.L.S., J.I.R., A.H., P.M.R., B.M.P., J.C.M.W., A.G.U., Y.L., B.H.C.S., S.B.F., S.R.H., E.J.B., V.G. and S.K. P.T.E., K.L.L., C.M.A., N.L.G., M.D.R., A.V.S., D.E.A., M.M.-N., B.P.K., N.A.L., J.C.B., M.K.C., M.D., K.O., T. Tanaka, B.H.C.S., S.B.F., A.A., D.D., J.B., D.I.C., S.R.H., V.G., E.J.B. and S. Kääb had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Fuster V, et al. ACC/AHA/ESC 2006 Guidelines for the Management of Patients with Atrial Fibrillation: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines and the European Society of Cardiology Committee for Practice Guidelines (Writing Committee to Revise the 2001 Guidelines for the Management of Patients With Atrial Fibrillation): developed in collaboration with the European Heart Rhythm Association and the Heart Rhythm Society. Circulation. 2006;114:e257–e354. doi: 10.1161/CIRCULATIONAHA.106.177292. [DOI] [PubMed] [Google Scholar]
- 2.Ellinor PT, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat. Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin EJ, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat. Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudbjartsson DF, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat. Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfeufer A, et al. Genome-wide association study of PR interval. Nat. Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 8.Lubitz SA, et al. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. J. Am. Med. Assoc. 2010;304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergwerff M, et al. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- 10.Ihida-Stansbury K, et al. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ. Res. 2004;94:1507–1514. doi: 10.1161/01.RES.0000130656.72424.20. [DOI] [PubMed] [Google Scholar]
- 11.Volonte D, McTiernan CF, Drab M, Kasper M, Galbiati F. Caveolin-3 form heterooligomeric complexes in atrial cardiac myocytes that are required for doxorubicin-induced apoptosis. Am. J. Physiol. Heart Circ. Physiol. 2008;294:H392–H401. doi: 10.1152/ajpheart.01039.2007. [DOI] [PubMed] [Google Scholar]
- 12.Zhao YY, et al. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc. Natl. Acad. Sci. USA. 2002;99:11375–11380. doi: 10.1073/pnas.172360799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, et al. The regulation of the cardiac potassium channel (HERG) by caveolin-1. Biochem. Cell Biol. 2008;86:405–415. doi: 10.1139/o08-118. [DOI] [PubMed] [Google Scholar]
- 14.Sinner MF, et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG) Eur. Heart J. 2008;29:907–914. doi: 10.1093/eurheartj/ehm619. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum. Mol. Genet. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 16.Matsuura T, et al. Two newly identified genomic mutations in a Japanese female patient with fructose-1,6-bisphosphatase (FBPase) deficiency. Mol. Genet. Metab. 2002;76:207–210. doi: 10.1016/s1096-7192(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 17.Schulze-Bahr E, et al. Pacemaker channel dysfunction in a patient with sinus node disease. J. Clin. Invest. 2003;111:1537–1545. doi: 10.1172/JCI16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N. Engl. J. Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 19.Frey N, Olson EN. Calsarcin-3, a novel skeletal muscle–specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 20.Beqqali A, et al. CHAP is a newly identified Z-disc protein essential for heart and skeletal muscle function. J. Cell Sci. 2010;123:1141–1150. doi: 10.1242/jcs.063859. [DOI] [PubMed] [Google Scholar]
- 21.Arola AM, et al. Mutations in PDLIM3 and MYOZ1 encoding myocyte Z line proteins are infrequently found in idiopathic dilated cardiomyopathy. Mol. Genet. Metab. 2007;90:435–440. doi: 10.1016/j.ymgme.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 22.Brugada R, et al. Identification of a genetic locus for familial atrial fibrillation. N. Engl. J. Med. 1997;336:905–911. doi: 10.1056/NEJM199703273361302. [DOI] [PubMed] [Google Scholar]
- 23.Lubitz SA, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–984. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kääb S, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur. Heart J. 2009;30:813–819. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stranger BE, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasan RS, et al. Genetic variants associated with cardiac structure and function: a meta-analysis and replication of genome-wide association data. J. Am. Med. Assoc. 2009;302:168–178. doi: 10.1001/jama.2009.978-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson AD, et al. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.