Figure 1. Workflow and results of kinase substrate motif assay.
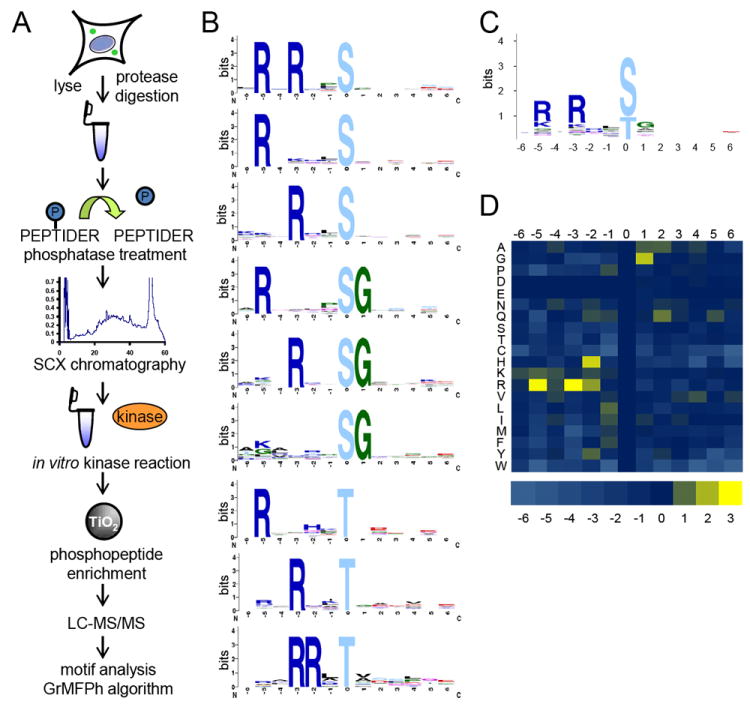
(A) Depiction of peptide kinase assay workflow. HeLa cells were lysed, digested, dephosphorylated, and separated into 12 fractions by strong cation exchange (SCX) chromatography. Select fractions were used in in vitro kinase reactions. Phosphopeptides were purified from non-phosphopeptides using titanium dioxide (TiO2) microspheres and analyzed by LC-MS/MS. Statistically significant motifs were extracted from the identified phosphopeptides by the in-house developed motif algorithm GrMPh. (B) Linear kinase motifs from Pim1 in vitro kinase reaction. (C) Averaged motif of all motif-containing peptides in (B). (D) Heat map representation of the log2 values of the ratio of foreground to background amino acid frequencies of motif-containing peptides in (A). See also Figures S1 – S9, S11 – S15, and Tables S1 and S2.
