Abstract
Disruption of the Pex2 gene leads to peroxisome deficiency and widespread metabolic dysfunction. We previously demonstrated that peroxisomes are critical for maintaining cholesterol homeostasis, using peroxisome-deficient Pex2−/− mice on a hybrid Swiss Webster×129S6/SvEv (SW/129) genetic background. Peroxisome deficiency activates hepatic endoplasmic reticulum (ER) stress pathways, leading to dysregulation of the endogenous sterol response mechanism. Herein, we demonstrate a more profound dysregulation of cholesterol homeostasis in newborn Pex2−/− mice congenic on a 129S6/SvEv (129) genetic background, and substantial differences between newborn versus postnatal Pex2−/− mice in factors that activate ER stress. These differences extend to relationships between activation of genes regulated by SREBP-2 versus PPARα. The SREBP-2 pathway is induced in neonatal Pex2−/− livers from 129 and SW/129 strains, despite normal hepatic cholesterol levels. ER stress markers are increased in newborn 129 Pex2−/− livers, which occurs in the absence of hepatic steatosis or accumulation of peroxins in the ER. Moreover, the induction of SREBP-2 and ER stress pathways is independent of PPARα activation in livers of newborn 129 and SW/129 Pex2−/− mice. Two-week-old wild-type mice treated with the peroxisome proliferator WY-14,643 show strong induction of PPARα-regulated genes and decreased expression of SREBP-2 and its target genes, further demonstrating that SREBP-2 pathway induction is not dependent on PPARα activation. Lastly, there is no activation of either SREBP-2 or ER stress pathways in kidney and lung of newborn Pex2−/− mice, suggesting a parallel induction of these pathways in peroxisome-deficient mice. These findings establish novel associations between SREBP-2, ER stress and PPARα pathway inductions.
Keywords: Cholesterol homeostasis, peroxisomes, Pex2, SREBP-2, ER stress, PPARα
1. Introduction
Peroxisomes are ubiquitous and highly versatile organelles of eukaryotic cells that have many metabolic functions, including β-oxidation and α-oxidation of fatty acids, ether-phospholipid synthesis, cholesterol and isoprenoid metabolism, bile acid synthesis, and metabolism of reactive oxygen species [1, 2, 3]. The importance of peroxisomes for normal cellular functioning is illustrated by the disorders of the Zellweger spectrum (Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum’s disease) in which functional peroxisomes are deficient [4].
Cholesterol is an important component of many cellular membranes, in particular the plasma membrane, and an obligatory precursor for synthesis of steroid hormones, bile acids, and regulatory oxysterols [5, 6]. The synthesis of cholesterol is a multi-step process involving nearly 30 enzymes. The pre-squalene segment of the cholesterol biosynthetic pathway is localized to peroxisomes, and acetyl-CoA derived from peroxisomal β-oxidation of very long-chain fatty acids (VLCFAs) and dicarboxylic acids is channeled preferentially to cholesterol synthesis inside the peroxisomes [7].
Cellular cholesterol levels are tightly regulated and reflect a delicate balance between dietary uptake, efflux, endogenous synthesis, and conversion of cholesterol to bile acids [5, 6, 8]. Cells contain an elaborate feedback system that senses cholesterol levels and modulates the transcription of genes that mediate cholesterol synthesis and uptake. Central to the regulatory system is the sterol regulatory element-binding protein (SREBP) family of transcription factors [8]. Accessory proteins [SREBP cleavage-activating protein (SCAP), insulin-induced genes 1 and 2 (Insig-1 and -2), and proteases (Site-1 and Site-2 protease)] mediate the sensing of membrane composition and fluidity and the subsequent translocation and activation of the transcription factors [8, 9]. Fine-tuning of cholesterol biosynthesis via post-translational regulation of the rate-limiting enzyme 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase (HMGCR) is achieved through Insig-1-dependent proteasomal degradation, which also responds to cholesterol levels in the ER [8].
The PEX2 protein (Pex2p) is a peroxisomal integral membrane protein involved in the import of peroxisomal matrix proteins; its absence in both patients with peroxisomal defects and Pex2−/− mice results in a lack of functional peroxisomes and abnormal peroxisomal biochemical parameters (i.e., increased levels of VLCFAs, a deficiency in plasmalogens, and localization of catalase to the cytosol) [10]. The Pex2 null allele has been bred on several different mouse genetic backgrounds, which markedly affects the survival of Pex2−/− mice. Homozygous Pex2−/− mice on a hybrid C57BL/6×129SvJ genetic background usually die on the day of birth (P0) [10], whereas Pex2−/− mice on a Swiss Webster×129S6/SvEv genetic background (SW/129) survive one to three weeks (rarely 5 weeks) [11, 12, 13]. When the Pex2 null allele is congenic on either a 129S6/SvEv (129), C57BL/6 or Swiss Webster genetic background, there is significant loss of homozygous mutants during embryogenesis, with only 20–50% surviving to birth and all mutants invariably dying on the day of birth [11]. Clearly there are as yet undefined genetic modifiers that affect the severity of the Pex2−/− phenotype.
Recent studies in postnatal SW/129 Pex2−/− mice have defined the important role of peroxisomes in maintaining normal cholesterol homeostasis [14, 15]. Despite an increased rate of hepatic cholesterol biosynthesis and activation of SREBP-2 target genes involved in cholesterol biosynthesis, early postnatal Pex2−/− mice have reduced cholesterol levels in both plasma and liver. While oral bile acid treatment normalized hepatic and plasma cholesterol levels and hepatic cholesterol synthesis in early postnatal Pex2−/− mice, SREBP-2 and its target gene expressions remained increased [15], suggesting an additional cholesterol-independent regulatory mechanism controlling the SREBP-2 pathway. This induction was also observed in liver of P0 and P36 SW/129 Pex2−/− mice, despite normal hepatic cholesterol levels [15]. We showed that peroxisome deficiency activates hepatic endoplasmic reticulum (ER) stress pathways, especially the integrated stress response (ISR) mediated by PERK (PKR-like endoplasmic reticulum kinase) and ATF4 (activating transcription factor-4) signaling, and hypothesized that ER stress leads to dysregulation of the endogenous sterol response mechanism and SREBP-2 pathway induction [15].
Several studies have suggested an involvement of the peroxisome proliferator-activated receptor alpha (PPARα) in the regulation of cholesterol synthesis; however, both stimulatory and inhibitory effects of PPARα have been reported [16–21]. PPARα pathways are up-regulated in peroxisome-deficiency and when rodents are treated with drugs that cause peroxisome proliferation (e.g., WY-14,643) [22, 23, 24]. ER stress also occurs in disorders associated with fatty liver [25], and Pex2−/− mice develop steatosis in the postnatal period [13]. In the present study, we characterize the regulation of cholesterol homeostasis and ER stress pathways in P0 Pex2−/− mice from both 129 and SW/129 strains, and evaluate the relationship between activation of genes regulated by SREBP-2 versus PPARα. We found that the induction of ER stress pathways occurs in the absence of hepatic steatosis or accumulation of peroxins (Pex proteins) in the ER in these newborn mutants. We present novel data to demonstrate that induction of the SREBP-2 pathway is independent of PPARα activation. In contrast to our findings in liver, organs that lack ER stress in newborn Pex2−/− mice also do not show SREBP-2 pathway induction, suggesting a parallel induction of these pathways in peroxisome-deficient mice.
2. Material and methods
2.1. Animals
Homozygous Pex2−/− mice were obtained by breeding Pex2 heterozygotes on a congenic 129S6/SvEv background or a hybrid Swiss Webster-129 (SW/129) background [11]. Mice had access to food and water ad libitum and were exposed to a 12-hour light-dark cycle. For the purposes of this study, control mice consisted of either Pex2+/+ (wild-type) or Pex2+/− genotypes, as their biochemical characteristics were comparable to one another [14].
2-week-old mice on a mixed genetic background (129Sv/J, C57BL/6J) received a single daily gavage dose of either WY-14,643 (50mg/kg/day; Enzo Life Sciences) suspended in methylcellulose (0.1%) or the carrier methylcellulose alone for 7 days [23].
All protocols for animal use and experiments were reviewed and approved by the Institutional Animal Care and Use Committee of San Diego State University and of Columbia University and by the Veterinary Office of Zürich (Switzerland).
2.2. Plasma and hepatic lipid analysis
Tissue sterols and plasma total cholesterol, HDL and LDL cholesterol, phospholipids, and triglycerides were measured as described previously [14].
2.3. Enzyme assays
3-hydroxy-3-methylglutaryl (HMG)-CoA reductase (HMGCR; EC 1.1.1.34), farnesylpyrophosphate synthase (FPPS; EC 2.5.1.1), isopentenyldiphosphate isomerase (IDI1; EC 5.3.3.2), squalene synthase (SQS; EC 2.5.1.21), and catalase activities were assayed as described previously [14]. Protein concentration was determined by the BCA method (Pierce, Rockford, IL, USA).
2.4. Western blot analysis
Proteins were separated on SDS-polyacrylamide gels. Immunoblot analysis was performed by enhanced chemiluminescence (PerkinElmer Life Sciences, Boston, MA, USA) with the following antibodies: anti-HMGCR, anti-IDI1, anti-FPPS (a gift of P. Edwards, UCLA), anti-(acyl-CoA oxidase 1) (a gift of Dr. A. Voelkl, University of Heidelberg), anti-ADFP (Abcam, Cambridge, MA, USA), anti-Pex14p (ProteinTech Group, Chicago, IL, USA), anti-Pex3p (ProteinTech Group), anti-Pex16p (ProteinTech Group), anti-catalase (Calbiochem, Nottingham, UK), anti-actin (Sigma-Aldrich, Buchs, Switzerland), anti-phospho-eIF2α (Ser51) (Cell Signaling, Danvers, MA, USA), anti-phospho-HMGCR (Ser872) (Millipore, Billerica, MA, USA), anti-phospho-AMPKα (Thr172) (Cell Signaling, Danvers, MA, USA), anti-AMPKα (Cell Signaling) with the appropriate horseradish peroxidase-linked secondary antibody (Bio-Rad, Hercules, CA). Blots were exposed to Kodak X-OMAT LS film (Rochester, NY), scanned on a densitometer (Molecular Dynamics) and analyzed with ImageQuant® software (Amersham Pharmacia Biotech, Piscataway, NJ).
2.5. Quantitative real-time RT-PCR
Total RNA was prepared from frozen mouse tissues with RNeasy Mini Kit (QIAGEN, Hilden, Germany) and treated with DNase I (DNA-free; Ambion, Austin, TX). First-strand cDNA was synthesized with random hexamer primers using Ready-To-Go You-Prime First-Strand Beads (Amersham Biosciences, Freiburg, Germany). The real-time RT-PCR reaction was set up in a final volume of 20 μl using 2× LightCycler 480 SYBR Green I Mastermix (Roche Diagnostics, Mannheim, Germany). PCR reactions were performed in triplicate using a Roche LightCycler 480. Relative mRNA amount was calculated using the comparative threshold cycle (CT) method. 18S rRNA was used as the invariant control. Primer sequences are available on request. Northern blot analysis was performed as described previously [14].
2.6. Histology and immunohistochemistry
Mice were cardiac-perfused with 4% paraformaldehyde (PFA)-PBS. The liver was post-fixed overnight in PFA-PBS and either cryoprotected in 30% sucrose-PBS or processed for paraffin embedding. For histologic detection of lipids, 12-μm thick cryostat sections were mounted on Superfrost Plus slides (Fisher Scientific) and stained with Oil Red O in propylene glycol [26]. For immunohistochemistry, 2-μm thick paraffin-embedded liver sections were mounted on Superfrost Plus slides; for antigen retrieval, sections were digested with 0.01% trypsin for 10 min at 37 °C followed by microwaving in 10 mM citrate buffer (pH 6.0) for 3 × 5 min at 800 W. Nonspecific binding was blocked with 4% BSA and 0.05% Tween 20 in PBS for 2 h and sections were then incubated overnight at 4 °C with rabbit anti-Pex14p, rabbit anti-Pex3p, rabbit anti-Pex16p or anti-catalase. Alexa Fluor 488-conjugated goat anti-rabbit IgG was applied for 2 h (Invitrogen, Carlsbad, CA). In addition, to detect Pex3p and Pex16p in liver sections we increased the sensitivity of the fluorescent stain by using an avidin-biotin system for amplification of the label (Vector Laboratories, Burlingame, CA). Sections were incubated for 2 h with biotin-conjugated anti-rabbit IgG (Jackson ImmunoResearch, Suffolk, UK) and subsequently with Fluorescein Avidin DCS for 20 min (Vector Laboratories). Negative control sections were incubated in parallel by omitting the primary antibody. Images were taken with a Leica TCS-SP1 confocal laser scanning microscope.
2.7. Statistical Analyses
Data are expressed as means ± S.D. Statistical significance was evaluated by an unpaired Student’s t-test.
3. Results
3.1. Plasma lipid and tissue cholesterol analysis of control and 129 Pex2−/− mice in comparison to SW/129 Pex2−/− mice
We first investigated if newborn peroxisome-deficient mice from 129 and SW/129 strains had abnormalities in plasma lipids (Table 1). Compared to controls, total plasma cholesterol was reduced by 40% and 33% in 129 and SW/129 Pex2−/− mice, respectively; HDL cholesterol was reduced by 45% in P0 129 Pex2−/− mice, but did not differ from controls in P0 SW/129 Pex2−/− mice. The plasma phospholipid level was similar in controls from both strains; it was reduced by 38% in SW/129 Pex2−/− mice and even more severely reduced by 61% in 129 Pex2−/− mice. In 129 strain mice, the calculated plasma LDL concentration was reduced by 46% in Pex2 mutants, whereas plasma triglycerides varied widely and did not differ significantly between controls and Pex2−/− mice. In comparison with early postnatal (P10) SW/129 Pex2−/− mice [14], the extent of reduction in total plasma cholesterol level is highly similar; however, plasma HDL and LDL cholesterol and phospholipids are more severely reduced in P0 129 mutants. Total cholesterol levels in the liver (2.87 ± 0.22 mg/g wet weight), kidney (2.76 ± 0.27 mg/g), and brain (3.6 ± 0.14 mg/g) were similar in P0 129 control and Pex2−/− mice.
Table 1.
Plasma lipid analysis of newborn control and Pex2−/− mice on the 129 and SW/129 genetic background
| Lipid | Concentration (mg/dl)
|
|||
|---|---|---|---|---|
| 129 | SW/129 | |||
| Control mice | Pex2−/− mice | Control mice | Pex2−/− mice | |
| Total plasma cholesterol | 58.8 ± 10.0 (32) | 35.4 ± 9.2 (11) ** | 67.1 ± 9.9 (17) | 45.0 ± 12.7 (6) ** |
| Plasma HDL cholesterol | 18.4 ± 10.1 (23) | 10.1 ± 6.1 (11) * | 18.0 ± 5.8 (16) | 17.8 ± 4.8 (7) |
| Plasma triglycerides | 62.8 ± 35.8 (16) | 44.4 ± 35.7 (8) | ND | ND |
| Plasma phospholipids | 150.6 ± 23.1 (18) | 59.3 ± 20.1 (7) ** | 163.4 ± 22.9 (17) | 100.5 ± 18.5 (5)** |
| Plasma LDL cholesterol | 32.3 | 17.6 | ND | ND |
Each value represents the mean ± S.D. Plasma lipid concentrations from Pex2 wild-type and heterozygous mice were similar and were combined (control mice). Values in parentheses denote the number of samples analyzed. Plasma LDL cholesterol was calculated according to the Friedewald formula applying the values of total cholesterol, HDL cholesterol, and triglycerides.
, P < 0.05 and
, P < 0.001 denote the level of statistical significance (Student’s t-test). ND, not determined.
3.2. Cholesterol biosynthetic enzyme activities and protein levels
We previously reported [14] that the activities of several cholesterol biosynthetic enzymes, including HMG-CoA reductase (HMGCR), IPP isomerase (IDI1), FPP synthase (FPPS) and squalene synthase (SQS), were normal in livers of newborn SW/129 strain Pex2−/− mice compared to age-matched controls, but all enzyme activities were significantly elevated in the postnatal knockout mouse livers. In contrast, activities of HMGCR and SQS were already increased 2.1-fold, and IDI1 activity was increased 1.6-fold in the liver of P0 129 Pex2−/− mice, relative to controls; the activity of FPPS was similar in the liver of control and 129 Pex2−/− mice (cf. Table 2 and [14]).
Table 2.
Activities of cholesterol biosynthetic enzymes in liver and kidney of P0 control and Pex2−/− mice on the 129 and SW/129 genetic background
| Enzyme | Liver
|
|||
|---|---|---|---|---|
| 129 | SW/129 | |||
| Control | Pex2−/− | Control | Pex2−/− | |
| HMG-CoA reductase | 254.3 ± 109.0 (12) | 532.8 ± 127.5 (6) *** | 189.9 ± 48.6 (4) | 215.7 ± 21.6 (3) |
| IPP isomerase | 2030.1 ± 541.4 (10) | 3148.9 ± 648.4 (6) ** | 944.5 ± 154.9 (4) | 1250.5 ± 125.0 (3) |
| FPP synthase | 3051.2 ± 411.7 (10) | 3328.1 ± 240.3 (7) | 3400.4 ± 388.6 (4) | 3768.5 ± 505.5 (3) |
| Squalene synthase | 8.6 ± 4.4 (8) | 17.6 ± 4.8 (5) ** | 11.9 ± 3.0 (4) | 22.3 ± 4.0 (3) |
| Enzyme | Kidney
|
|||
| 129 | SW/129 | |||
| Control | Pex2−/− | Control | Pex2−/− | |
| HMG-CoA reductase | 64.2 ± 21.5 (5) | 33.1 ± 6.0 (5) * | 81.1 ± 19.5 (6) | 42.1 ± 8.7 (5) *** |
| IPP isomerase | 421.0 ± 40.8 (3) | 396.7 ± 83.8 (3) | 447.1 ± 27.3 (6) | 444.9 ± 59.7 (5) |
| FPP synthase | 587.5 ± 37.4 (7) | 524.8 ± 53.9 (5) | 624.7 ± 81.4 (6) | 549.0 ± 76.3 (5) |
| Squalene synthase | 2.01 ± 0.74 (7) | 1.30 ± 0.44 (5) | 4.56 ± 0.31 (3) | 3.77 ± 0.27 (3) * |
Each value represents the mean ± S.D. of the activities expressed as pmol/min/mg protein (n = number of samples). Enzyme activities from wild-type and heterozygous mice were similar and combined (control mice).
, P < 0.05;
, P < 0.005;
P < 0.001 denote the level of statistical significance (Student’s t-test). Activities of cholesterol biosynthetic enzymes in liver of P0 mice on the SW/129 genetic background are reproduced, with permission, from Kovacs et al. [14], © American Society for Microbiology [Mol. Cell. Biol., 2004; 24: 1–13.]
While HMGCR activity was highly increased in livers of postnatal SW/129 Pex2−/− mice compared to control mice, its activity was decreased by ~40% in kidneys of postnatal Pex2 mutants [14, 15]. Similarly, the activity of HMGCR was decreased by 48% in kidneys of P0 129 and SW/129 Pex2−/− mice compared to controls (Table 2). This appears to be a posttranscriptional regulation, as the mRNA levels of HMGCR were similar in P0 control and Pex2−/− mice (Fig. 8A). In contrast, activities of IDI1, FPPS, and SQS were similar in kidneys of P0 control and Pex2−/− mice (Table 2).
Figure 8. Expression of ER stress markers and SREBP-2-regulated genes in kidney and lung from P0 129 control and Pex2−/− mice.
(A) Kidney (n = 4 for control and Pex2−/− mice). (B) Lung (n = 6 for control mice; n = 4 for Pex2−/− mice). RNA was analyzed by quantitative RT-PCR. Each value represents the amount of mRNA relative to that in control mice, which was arbitrarily defined as 1. Values are mean ± S.D. from RNA samples of individual mice. *, P < 0.05; **, P < 0.01 vs. control mice (Student’s t-test).
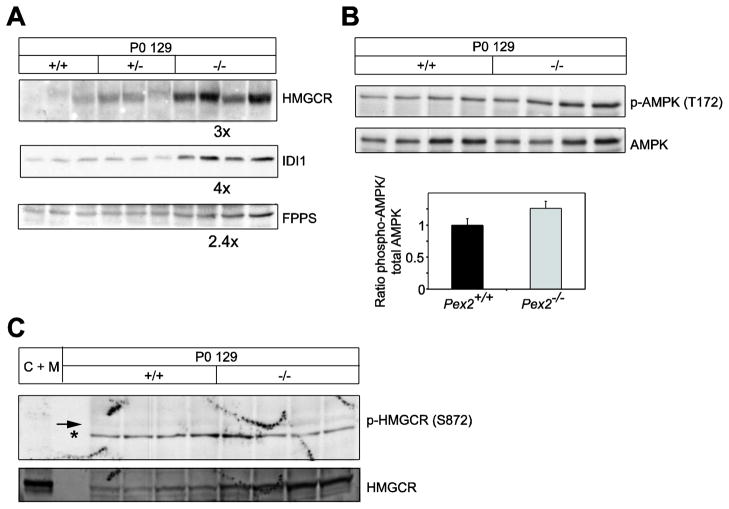
Western blot analysis of cholesterol biosynthetic enzymes was performed to determine whether the measured activities were a reflection of the protein levels. HMGCR protein levels were increased 3-fold, and IDI1 and FPPS protein levels were elevated 4-fold and 2.4-fold, respectively, in 129 Pex2−/− liver compared to controls (Fig. 1A). In comparison, similar protein levels of most cholesterol biosynthetic enzymes were seen in P0 SW/129 control and Pex2−/− livers [14].
Figure 1. Immunoblot analysis of cholesterol biosynthetic enzymes, AMPK activation state, and HMGCR phosphorylation state in extracts of livers of individual control and Pex2−/− mice.
Whole liver lysates were subjected to immunoblot analysis with the indicated antibodies. (A) Values for fold change of protein levels in Pex2−/− mice (shown below each corresponding blot) are expressed relative to that in control mice (wild-type and heterozygous), which in each case is given a value of 1. (B) The AMPK activation state is expressed as the ratio between the densitometric analysis of phospho-AMPK and total AMPK. Ratios from control mice were arbitrarily set at 1. (C) Immunoblot analysis of phospho-HMGCR and total HMGCR. As positive control 30 μg protein of liver homogenate from a cholestyramine plus mevinolin-treated rat, a treatment known to significantly induce cholesterol biosynthetic proteins, was used (C + M). An arrow points to phosphorylated HMGCR (p-HMGCR); nonspecific bands are denoted by the asterisk. Note that phosphorylation of HMGCR at Ser872 is very weak and similar in control and Pex2−/− livers, whereas total HMGCR protein levels were significantly increased in Pex2−/− livers.
We previously demonstrated that an estimated “catalytic efficiency” of HMGCR and IDI1, obtained by normalizing the enzyme specific activities to the enzyme protein content within each liver homogenate, was significantly decreased both in untreated and bile acid-fed postnatal SW/129 Pex2−/− mice, relative to controls [14, 15]. In P0 129 Pex2−/− liver, the catalytic efficiencies of HMGCR, IDI1, and FPPS were decreased by ~40% (P=0.05) (supplemental Table S1), ~60% (P<0.001) (supplemental Table S2), and ~50% (P=0.06) (data not shown), respectively.
In addition to controlling HMGCR activity by regulating its transcription and protein degradation, mammalian cells also modulate HMGCR activity by phosphorylation [27]. A high AMP:ATP ratio activates the AMP-activated protein kinase (AMPK), which phosphorylates a conserved serine (Ser872) in the HMGCR active site and thereby decreases HMGCR activity. To determine whether the decreased efficiency of HMGCR in Pex2−/− liver is due to AMPK-directed HMGCR phosphorylation, hepatic AMPK activity was assessed by determining the phosphorylation state of AMPK. Hepatic phosphorylation of AMPK at Thr172 was not significantly altered in P0 Pex2−/− liver compared to control liver (Fig. 1B). Furthermore, phosphorylation of HMGCR at Ser872 was very weak and similar in control and Pex2−/− livers (Fig. 1C).
In summary, the protein level and enzyme activities of hepatic cholesterol biosynthetic enzymes are more severely altered in P0 Pex2−/− mice from 129 strain versus that in SW/129 strain, but HMGCR activity is not differentially modulated by phosphorylation in newborn control versus Pex2 mutant liver.
3.3. Expression of cholesterol biosynthetic genes in the liver of 129 Pex2−/− mice
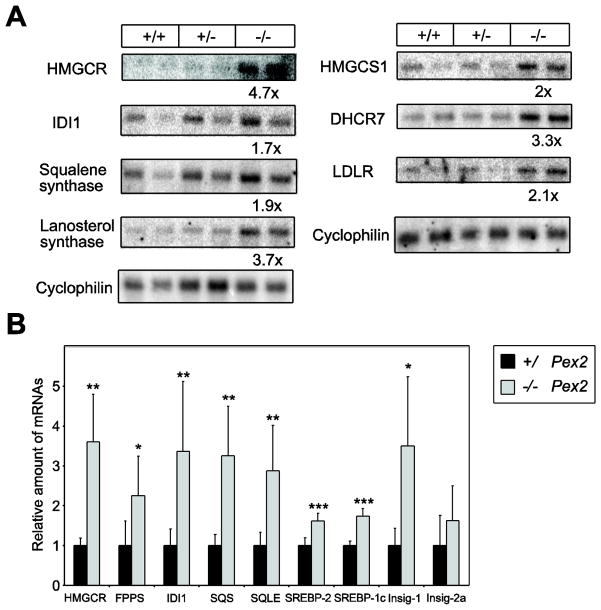
We previously reported that the hepatic expression of genes encoding SREBP-2-regulated cholesterol biosynthetic enzymes was markedly increased in postnatal SW/129 Pex2−/− mice compared to control mice, which occurs in association with reduced hepatic and plasma total cholesterol levels in mutant mice [14, 15]. However, as hepatic cholesterol levels are similar in P0 129 Pex2−/− and control mice, we examined whether SREBP pathway genes were affected in these mutants (Fig. 2A, B). The expression of SREBP-2 and its target genes HMG-CoA synthase 1 (HMGCS1), HMGCR, FPPS, IDI1, SQS, squalene epoxidase (SQLE), lanosterol synthase (LSS), 7-dehydrocholesterol reductase (DHCR7), LDL receptor (LDLR) and Insig-1 were all significantly increased 1.6- to 4.7-fold in P0 129 Pex2−/− versus control mouse liver. Insig-2a expression levels were similar in 129 control and Pex2−/− liver. SREBP-1c expression was significantly increased 1.7-fold in P0 129 Pex2−/− versus control liver (Fig. 2B), which is opposite to the significantly reduced hepatic SREBP-1c expression in P0, P10, and P36 SW/129 Pex2−/− mice [14, 15].
Figure 2. Northern blot (A) and quantitative RT-PCR analysis (B-E) of SREBP-regulated genes in livers from P0 129 control and Pex2−/− mice.
The values obtained were normalized either to cyclophilin (A) or 18S rRNA (B) values. (A) Aliquots (3 μg) of mRNA (equal aliquots of RNA from three mice were pooled) were subjected to electrophoresis and blot hybridization with the indicated 32P-labeled probe. The amount of radioactivity in each band was quantified by PhosphorImager analysis and normalized to the signal generated by cyclophilin. The fold change in each mRNA of Pex2−/− mice is expressed relative to control mice, which in each case was arbitrarily set at 1. (B) Each value represents the amount of mRNA relative to that in control mice, which was arbitrarily defined as 1. Values are mean ± S.D. from RNA samples of six individual mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. control mice (Student’s t-test).
In summary, there is a very similar mRNA up-regulation of SREBP-2 and its target genes in livers of newborn Pex2−/− mice from both 129 and SW/129 strains (supplemental Table S3).
3.4. SREBP-2 dysregulation is independent of PPARα activation in the liver of newborn 129 and SW/129 Pex2−/− mice
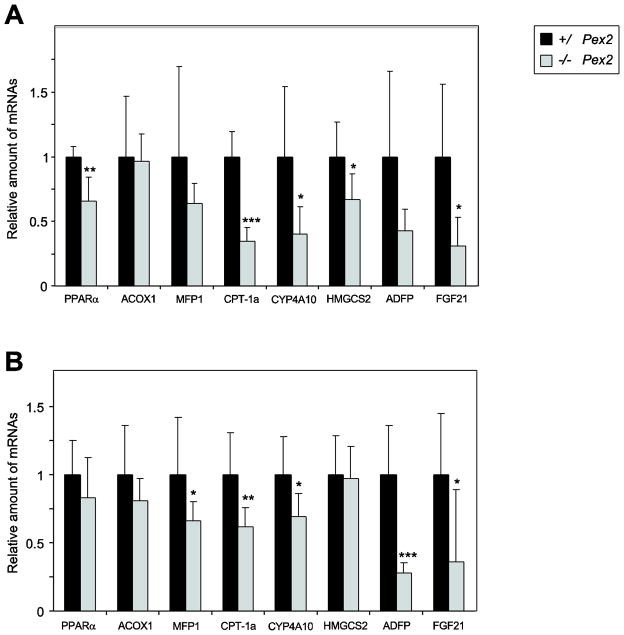
Recently, it has been suggested that induction of PPARα leads to upregulation of the SREBP-2 pathway [28]. Targets of PPARα, a nuclear receptor involved in the regulation of energy homeostasis, are induced in postnatal peroxisome-deficient SW/129 Pex2−/− livers, probably as a result of increased concentrations of endogenous PPARα lipid ligands (W.J. Kovacs and P.L. Faust, unpublished results). To investigate whether the PPARα signaling pathway was activated in liver of newborn 129 Pex2−/− mice, mRNA levels of PPARα and several PPARα target genes were determined (Fig. 3A). The expression of the PPARα transcript was reduced by ~35% in 129 Pex2−/− mice compared to controls. The mRNA levels of the peroxisomal β-oxidation enzymes acyl-CoA oxidase 1 (ACOX1) and multifunctional protein 1 (MFP1) were similar in 129 control and Pex2−/− liver. The lipid droplet associated protein ADFP (adipose differentiation related protein) and fibroblast growth factor 21 (FGF21) are both direct target genes for PPARα in liver [29–33]. The mRNA expression of ADFP showed a tendency to decrease, and FGF21 was decreased by 70% in 129 Pex2−/− mice compared to controls. Transcript level of several other PPARα target genes was significantly decreased in 129 Pex2−/− versus control mouse liver, including carnitine palmitoyltransferase 1a (CPT-1a), which has a pivotal role in the regulation of mitochondrial β-oxidation, CYP4A10, a microsomal ω-oxidation enzyme, and mitochondrial HMG-CoA synthase (HMGCS2), which is a key control site of ketogenesis.
Figure 3. Quantitative RT-PCR analysis of various PPARα target genes in livers from P0 129 and SW/129 control andPex2−/− mice.
(A) 129 control and Pex2−/− mice (n = 6 for control and Pex2−/− mice). (B) SW/129 control and Pex2−/− mice (n = 10 for control mice; n = 9 for Pex2−/− mice). The values obtained were normalized to 18S rRNA values. Each value represents the amount of mRNA relative to that in control mice, which was arbitrarily defined as 1. Values are mean ± S.D. from RNA samples of individual mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. control mice (Student’s t-test).
Next, we investigated whether the PPARα signaling pathway was activated in newborn SW/129 Pex2−/− mice (Fig. 3B). Newborn SW/129 Pex2−/− mice, in contrast to P0 129 Pex2−/− mice, ingest food; hence, diet-derived biological ligands for PPARα might be present. The mRNA levels of MFP1, CPT-1a, and CYP4A10 were significantly decreased by ~30–40% in P0 SW/129 Pex2−/− mice (Fig. 3B). The expression of PPARα, ACOX1, and HMGCS2 was similar in SW/129 control and Pex2−/− mice (Fig. 3B). The mRNAs for ADFP and FGF21 were significantly decreased (70 and 65%, respectively) in SW/129 Pex2−/− mice relative to controls (Fig. 3B).
In summary, mRNA levels of PPARα target genes are either unchanged or decreased in both P0 129 and SW/129 Pex2−/− mice compared to controls.
3.5. PPARα activation does not induce the SREBP-2 pathway in the liver of wild-type postnatal mice
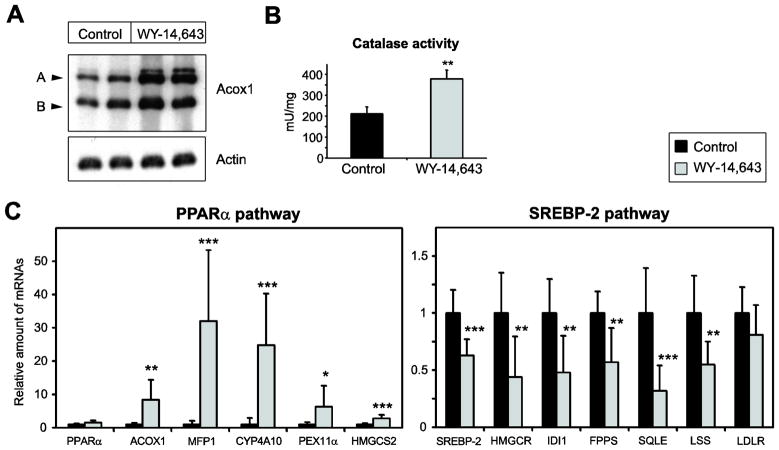
Since contradictory data regarding the role of PPARα activation on cholesterol synthesis have been reported, we determined the effect of PPARα activation on the SREBP-2 pathway in postnatal mice. Therefore, we treated 2-week-old wild-type mice with the PPARα activator WY-14,643 for 7 days. We chose to examine mice at this younger age, rather than adult mice, as our analyses of Pex2 mutants are also largely limited to this age group. A hallmark physiological response in rodent peroxisome proliferation is the induction of the peroxisomal β-oxidation enzymes. Protein levels of acyl-CoA oxidase 1 (Acox1) were strongly increased in livers of WY-14,643-treated mice (Fig. 4A). The activity of catalase, the marker enzyme of peroxisomes, was significantly increased 1.8-fold in WY-14,643-treated mice (Fig. 4B). Next, we determined the mRNA levels of several PPARα target genes in the liver of control and WY-14,643-treated mice. The mRNA levels of ACOX1, MFP1, CYP4A10, PEX11α, and HMGCS2 were significantly increased in WY-14,643-treated mice (Fig. 4C). The expression of the PPARα transcript was similar in control and WY-14,643-treated mice (Fig. 4C). The expression of SREBP-2 and its target genes HMGCR, IDI1, FPPS, SQLE, and LSS was significantly decreased in WY-14,643-treated vs. control mouse liver (Fig. 4D). The expression of the LDLR was similar in control and WY-14,643-treated mice (Fig. 4D).
Figure 4. Effect of PPARα activation on SREBP-2-regulated genes in the liver.
Mice were treated orally with 50 mg/kg of WY-14,643 for 7 days. Control mice received the appropriate volume of the vehicle. (A) Immunoblot analysis of acyl-CoA oxidase (Acox1) and actin in livers from control and WY-14,643-treated mice. Two subunits with molecular masses of 72 and 52 kDa (A and B, respectively) are shown. Note the strong increase of the protein levels of the PPARα target gene Acox1 in WY-14,643-treated mice. (B) Catalase activity in liver homogenates from control and WY-14,643-treated mice. Values are mean ± S.D. (n=8 for control and WY-14,643-treated mice). (C) Quantitative RT-PCR analysis of various PPARα and SREBP-2 target genes in livers from control and WY-14,643-treated mice. The values obtained were normalized to 18S rRNA values. Each value represents the amount of mRNA relative to that in control mice, which was arbitrarily defined as 1. Values are mean ± S.D. from RNA samples of eight individual mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. control mice (Student’s t-test).
3.6. Expression of ER stress markers in the liver of 129 Pex2−/− mice
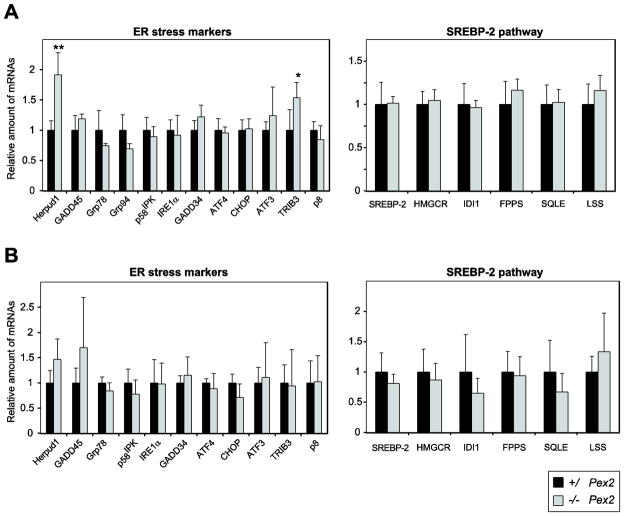
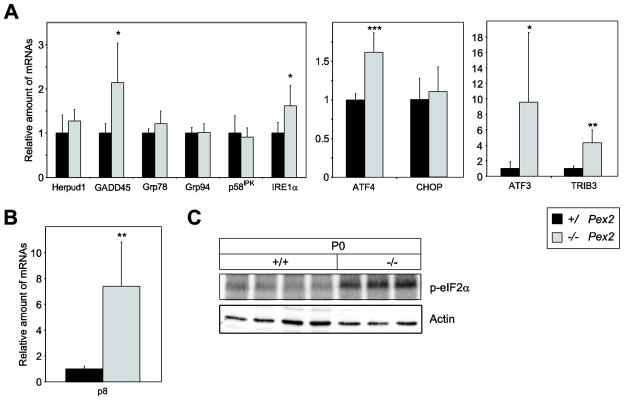
To determine whether ER stress is also present in P0 129 Pex2−/− livers, we examined the expression of several unfolded protein response (UPR) target genes. While the mRNA levels of ER chaperones glucose-regulated protein 78 (Grp78) and Grp94 and the co-chaperone protein p58IPK were similar in P0 129 control and Pex2−/− mice (Fig. 5A), the expression of GADD45 (growth arrest and DNA damage-inducible 45) was significantly increased 2.1-fold in 129 Pex2−/− mice (Fig. 5A). Herpud1 (homocysteine-inducible ER stress-inducible ubiquitin-like domain member 1) mRNA levels were similar in 129 control and Pex2−/− mice (Fig. 5A). IRE1α (inositol-requiring enzyme-1) is a dual function serine-threonine protein kinase and endoribonuclease. Activation of its RNAse domain results in the unconventional splicing of X-box binding protein 1 (XBP-1) mRNA and feedback downregulation of IRE1α mRNA [34]. The expression of IRE1α was significantly increased in 129 Pex2−/− liver (Fig. 5A). The expression levels of total and spliced XBP-1 were similar in P0 129 control and Pex2−/− mice (data not shown).
Figure 5. Expression of ER stress markers in livers from P0 129 control and Pex2−/− mice.
(A, B) RNA was analyzed by quantitative RT-PCR. The values obtained were normalized to 18S rRNA values. Each value represents the amount of mRNA relative to that in control mice, which was arbitrarily defined as 1. Values are mean ± S.D. from RNA samples of six individual mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001 vs. control mice (Student’s t-test). (C) Expression levels of phospho-eIF2α (p-eIF2α) and actin in livers from P0 129 control and Pex2−/− mice were measured by immunoblot.
The major determinant of ISR activity in liver is the ER stress-activated kinase PERK, whose transient activation is difficult to detect [35]. The best-characterized PERK substrate is the α subunit of the eukaryotic translation initiation factor 2 (eIF2α). Phosphorylation of eIF2α on serine 51 reduces protein translation and diminishes the load of unfolded proteins entering the ER [36]. eIF2α phosphorylation was significantly increased in P0 129 Pex2−/− liver (Fig. 5C). Phosphorylated eIF2α stimulates selective translation of the transcription factor ATF4 [37–39], which plays a crucial role for the adaptation to stress [40]. The expression of the ATF4 transcription factor was significantly increased in P0 129 Pex2−/− mice compared to controls (Fig. 5A). We determined the hepatic expression of ATF3, TRIB3 (tribbles homolog 3), and CHOP (C/EBP homologous protein), which are transcriptional targets of ATF4. ATF3 and TRIB3 mRNA levels were significantly increased in 129 Pex2−/− mice (Fig. 5A), whereas CHOP expression was similar in 129 control and Pex2−/− mice (Fig. 5A). The expression of the stress associated protein p8, a downstream target of PERK [41], was increased 7.5-fold in 129 Pex2−/− mice (Fig. 5B).
In summary, there are both similarities and differences in the hepatic stress responses between newborn 129 versus SW/129 Pex2−/− livers (supplemental Table S4). This comparison reveals strong similarities in induction of the ISR in peroxisome-deficient liver, but some differences in expression of Herpud1, GADD45, ATF4 and CHOP.
3.7. Peroxisomal membrane proteins do not accumulate in the liver of 129 and SW/129 Pex2−/− mice
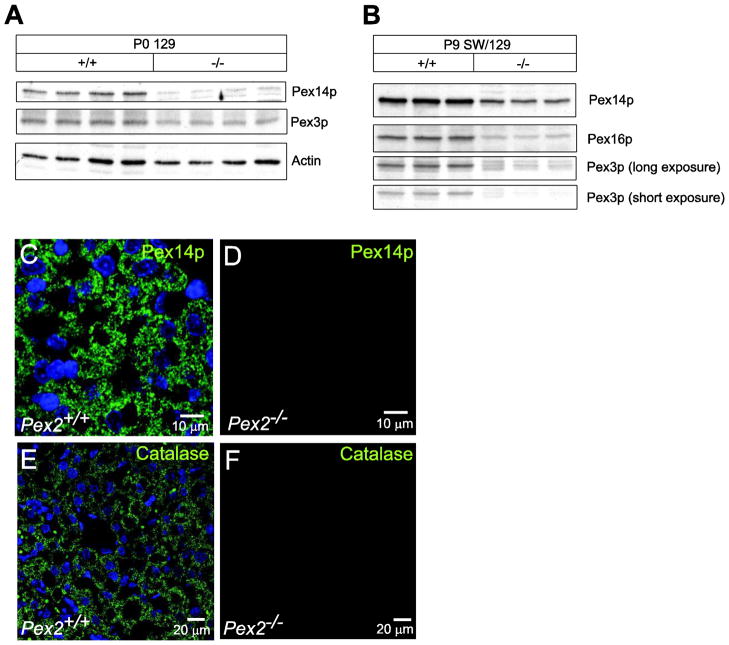
Proteins that pass through the ER on their way to the peroxisome might be retained in the ER and activate the unfolded protein response. A subset of peroxisomal membrane proteins is inserted into the ER for peroxisome biogenesis in yeast [42, 43], and co-translational insertion of overexpressed Pex16p and subsequent recruitment of Pex3p into the ER has been demonstrated in mammalian cells [44]. In Western blot analyses, the protein levels of Pex14p, Pex3p, and Pex16p were significantly decreased in the livers of newborn 129 and P9 SW/129 Pex2−/− mice compared to controls (Fig. 6A, B). This finding is most consistent with the small number of peroxisome membrane “ghosts” present in peroxisome-deficient cells [10], and suggests that peroxins (Pex proteins), which are proteins involved in peroxisome biogenesis and proliferation, do not accumulate in the ER and subsequently induce the UPR.
Figure 6. Immunoblot and immunofluorescence analysis of peroxin proteins and catalase in livers from control and Pex2−/− mice.
(A, B) Protein lysates from livers of newborn 129 (A) and 9-day-old SW/129 (B) control and Pex2−/− mice were probed by immunoblot as labeled. (C–F) Liver sections from P9 wild-type (C, E) and Pex2−/− (D, F) mice were stained with an antibody to Pex14p (C, D) and catalase (E, F) and imaged by confocal microscopy. Peroxisomes (C) and peroxisome membrane ghosts (D) were detected using an antibody to Pex14p. Note that the number of peroxisome membrane ghosts in Pex2−/− mice is significantly lower than the number of peroxisomes in wild-type mice. PTS1 protein import was assessed as the distribution of punctate (organelle-bound) (E) versus cytoplasmic (F) catalase. Scale bar: 10 μm for panels C, D; 20 μm for panels E, F.
To determine whether these peroxin proteins are localized in peroxisome membrane ghosts or retained in the ER, we examined the expression of Pex14p by immunohistochemistry in P9 wild-type and Pex2−/− livers (Fig. 6C–F); Pex3p and Pex16p could not be detected by immunohistochemistry in wild-type or Pex2−/− livers. A punctuate peroxisomal staining pattern for Pex14p was observed in liver sections from wild-type mice (Fig. 6C). In Pex2−/− livers, Pex14p was present at reduced levels in less abundant cellular vesicles, consistent with peroxisome membrane ghosts (Fig. 6D). We did not observe an ER localization of Pex14p in Pex2−/− livers. In addition, the immunofluorescence pattern obtained for catalase, a peroxisomal matrix protein, showed the characteristic punctuate peroxisomal distribution in wild-type liver cells (Fig. 6E) and a diffuse, cytoplasmic fluorescence in Pex2−/− livers (Fig. 6F), consistent with mislocalization of catalase to the cytoplasm. These findings are consistent with the established function of Pex2p in import of peroxisomal matrix, but not peroxisomal membrane proteins.
3.8. Newborn Pex2−/− mice do not display hepatic steatosis
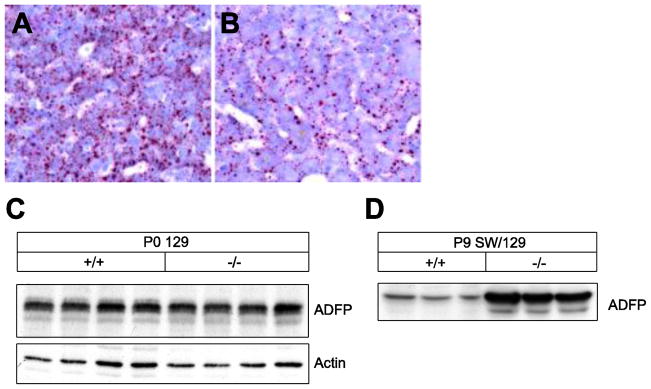
UPR activation has been observed in fatty liver diseases, suggesting the induction of ER stress in these pathological conditions [25]. Oil Red O staining shows a reduced content of neutral lipids in liver of P0 129 Pex2−/− versus control mice (Fig. 7A, B). ADFP localizes exclusively to lipid droplets, but is degraded by the ubiquitin-mediated proteasome pathway in the absence of excess neutral lipids [45–47]. ADFP protein levels were significantly increased in livers of early postnatal P9 SW/129 Pex2−/− mice (Fig. 7D); this correlates with the hepatic lipid accumulation in these mutants [13], which is predominantly present in triglycerides (data not shown). In contrast, the hepatic mRNA expression of ADFP was significantly decreased in P0 Pex2−/− mice (Fig. 3) and ADFP protein levels were similar in P0 129 control and Pex2−/− mice (Fig. 7C). These findings indicate that neutral lipid content is not significantly increased in newborn Pex2−/− liver and therefore does not contribute to the induction of hepatic ER stress in these mutants.
Figure 7. Analysis of lipid droplets in P0 129 Pex2−/− liver.
(A, B) Oil red O staining of liver cryostat sections from control (A) and Pex2−/− (B) mice shows a reduced number of lipid droplets in Pex2 mutants. (C, D) Protein levels of the lipid droplet-associated protein ADFP in livers of newborn 129 (C) and 9-day-old SW/129 (D) control and Pex2−/− mice were measured by immunoblot.
3.9. Absence of SREBP-2 or ER stress pathway induction in extrahepatic organs of newborn Pex2−/− mice
In this study, and in prior studies [15], we have demonstrated that induction of ER stress and SREBP-2 pathways occurs in parallel in liver of Pex2−/− mice. In contrast, there is no ER stress response in early postnatal kidney of Pex2−/− mice, where HMGCR activity is reduced, the activity of several other cholesterol enzymes is normal, and the rate of cholesterol biosynthesis is reduced [14, 15]. Furthermore, in both kidney and lung of P0 129 Pex2−/− mice, the mRNA expression level of ER stress and SREBP-2 related pathway genes was not increased compared to control mice (Fig. 8A, B). These findings support the observation that the induction of ER stress and cholesterol biosynthesis pathways occurs in parallel in peroxisome-deficient mice.
4. Discussion
Recently, we provided the first demonstration that peroxisome deficiency activates hepatic ER stress pathways, leading to dysregulation of the endogenous sterol response mechanism and subsequent SREBP-2 activation, which may occur despite normal hepatic cholesterol levels [15]. Here, we further extended these findings through a more detailed analyses of newborn (P0) Pex2−/− mice, and demonstrated that ER stress pathways are already induced in newborn peroxisome-deficient liver, and that this occurs in the absence of hepatic steatosis or accumulation of Pex proteins in the ER. In addition, mRNA levels of many PPARα target genes were reduced in the newborn Pex2 mutant liver, thus demonstrating that activation of SREBP-2 is not dependent on induction of PPARα pathways. Furthermore, treatment of early postnatal (2-week-old) wild-type mice with a peroxisome proliferator strongly induced hepatic PPARα target genes, but mRNA expression of SREBP-2 and several of its target genes was reduced. Expression analyses in extrahepatic organs of newborn Pex2−/− mice demonstrate that SREBP-2 and ER stress pathways are induced in parallel in peroxisome-deficient mice.
When the Pex2−/− allele is congenic on a 129 genetic background, we now have demonstrated that mutant mice have a more severe phenotype than those on a mixed SW/129 background, which is also reflected in the magnitude of the dysregulation in their cholesterol homeostasis. Our studies revealed that protein levels and activities of cholesterol biosynthetic enzymes are more severely altered in P0 Pex2−/− mice from 129 versus SW/129 strain, which again occurs despite normal cholesterol content in mutant livers. The increased mRNA expression of SREBP-2 and its target genes is similar in liver of newborn mutant mice on both 129 and SW/129 strains, consistent with our hypothesis that an up-regulation of the SREBP-2 pathway is necessary to maintain normal cholesterol levels in peroxisome-deficient mice. Despite the normal hepatic cholesterol level in all newborn Pex2−/− mice, plasma cholesterol levels are significantly reduced, indicating that SREBP-2 up-regulation in response to peroxisome deficiency may not be sufficient to maintain normal cholesterol homeostasis, as also seen in postnatal Pex2−/− mice [14]. The cholesterol biosynthetic pathway has also been analyzed in peroxisome-deficient Pex5−/− mice. Newborn Pex5−/− mice have normal levels of cholesterol in plasma, liver and brain [48] and the activity of cholesterol biosynthetic enzymes was either normal or slightly increased (HMGCR, IDI1) [49].
Recent studies have shown that cells can bypass the cholesterol inhibition of SREBP processing in response to ER stress and activate SREBP-2 [50–52]. Translation attenuation in ER-stressed cells due to PERK activation decreases levels of the protein Insig-1, thus releasing the cholesterol-sensing adaptor protein SCAP and SREBP-2 from inhibitory binding [51]. This leads to the translocation of SREBP-2 to the Golgi and generation of the active transcription factor. Interestingly, preliminary results show that the expression of SREBP-2 target genes (e.g., HMGCR, IDI1, FPPS, SQLE, LSS, SREBP-2) and ER stress markers (e.g., Grp78, TRIB3, ATF4, CHOP, p8) is already increased in livers from embryonic day 18.5 SW/129 Pex2−/− mice compared to controls (W.J. Kovacs and P.L. Faust, unpublished results). However, further studies are necessary to investigate the links between peroxisome deficiency, cholesterol biosynthesis, and UPR activation in fetal mice. To investigate if the activation of the SREBP-2 and ER stress pathways also go hand in hand in extrahepatic tissues, we determined the expression of SREBP-2 target genes and ER stress markers in the lung and kidney of P0 129 control and Pex2−/− mice. In contrast to liver, mRNA levels of SREBP-2 target genes and ER stress markers were similar in control and Pex2−/− lung and kidney (Fig. 8), suggesting that hepatic induction of SREBP-2 and its target genes is linked to the ER stress observed in the liver.
PPARα is a sensor for fatty acids and fatty acid derivatives, and thus controls important metabolic pathways involved in lipid and energy metabolism [53]. Potent endogenous PPARα ligands such as CoA thioesters of very-long chain and branched-chain fatty acids are metabolized in peroxisomes [53, 54], and accumulation of these unmetabolized substrates in postnatal peroxisome-deficient livers hyperactivates PPARα [W.J. Kovacs and P.L. Faust, unpublished results; 22]. Several studies suggested an involvement of PPARα in the regulation of cholesterol synthesis; however, both stimulatory and inhibitory effects of PPARα activators on hepatic HMGCR and cholesterol synthesis in rodents have been reported [16 – 21]. Here we show that the hepatic expression of PPARα and many PPARα target genes was either unchanged or significantly decreased in newborn 129 and SW/129 Pex2−/− mice (Fig. 3). Indeed, activation of PPARα-responsive genes in the liver of Acox1−/− mice does not occur during the embryonic period, but occurs as early as 1 day postnatal [55]. While newborn Pex2−/− mice from SW/129 strain do feed, the accumulation of abnormal metabolites may not yet be sufficient to induce the PPARα pathway. Thus, our findings in newborn Pex2−/− mice are consistent with the hypothesis that biological ligands for PPARα, such as VLCFAs or dicarboxylic acids, are absent in the developing embryo and are likely derived from the diet in the postnatal period.
The lack of PPARα target gene induction in P0 Pex2−/− mice does not support the hypothesis that induction of PPARα by endogenous ligands leads to upregulation of the SREBP-2 pathway. A tight interrelationship between induction of hepatic PPARα and SREBP-2 pathways was recently proposed in liver of 2-day-old and adult mice with inactivation of the D-specific multifunctional protein 2 (MFP2), which catalyzes the second and third step in peroxisomal β-oxidation [28]. However, newborn Mfp2−/− mice were not examined in their study. While sustained activation of PPARα in liver, either by synthetic or natural ligands, leads to ER and oxidative stress [53], this cannot account for the activation of ER stress in liver of newborn Pex2−/− mice.
The UPR is initiated by three ER membrane-associated proteins, IRE1α, PERK, and activating transcription factor (ATF)-6α [40, 56, 57, 58]. mRNA expression analysis of several UPR target genes revealed several similarities in the pattern of hepatic stress response in P0 129 versus P0 SW/129 Pex2−/− mice (supplemental Table S4), including increased expression of IRE1α (Fig. 5) and absence of XBP-1 mRNA splicing [15 and data not shown], suggesting that IRE1α signaling and its RNAse activity are not prominently induced in either newborn or postnatal Pex2−/− livers. All newborn Pex2−/− mice had normal hepatic expression of the ER chaperones Grp78 and Grp94 and the co-chaperone protein p58IPK, and prominent activation of the stress associated protein p8, a downstream target of PERK [41]. An opposite expression pattern was observed for Herpud1 and GADD45 in P0 129 versus SW/129 Pex2−/− mice. We previously noted that Herpud1 is initially upregulated in P0 SW/129 Pex2−/− liver, and its expression then decreases in postnatal mutant mice [15]. As Herpud1 is a component of the ER-associated protein degradation (ERAD) pathway that contributes to degradation of HMGCR protein [27], we may speculate that absence of ERAD up-regulation, now seen in P0 129 Pex2−/− liver, contributes to higher hepatic HMGCR protein levels in the 129 mutant Pex2 liver versus normal levels in SW/129 mutants. The integrated stress response further links PERK activation and subsequent ATF4 induction with other cellular adaptive pathways, including expression of phosphorylated eIF2α and ATF4 target genes, ATF3 and TRIB3 (Fig. 5). There is a consistent, strong induction of ISR related pathways in liver of postnatal Pex2−/− mice and in newborn mutants from both strains (Fig. 5, supplemental Table S4), indicating that peroxisome deficiency most prominently and consistently induces the ISR branch of the hepatic UPR. Recent studies demonstrate that all three isoforms of PPAR (α, γ, δ) interact with the p8 promoter to induce hepatic p8 gene expression, and sustained activation of PPARα pathways in Acox1−/− liver leads to ER stress [59]. As there is no activation of either PPARγ (data not shown) or PPARα in newborn Pex2−/− liver, clearly p8 is not an obligatory PPARα target for activation of p8 or ER stress in peroxisome-deficient mice. Our prior studies suggested that bile acids might activate p8 in Pex2−/− liver [15].
A still unresolved issue is how peroxisome dysfunction leads to the ER stress response. Because of the multiple abnormalities at the metabolite level in Pex2−/− mice it is probably impossible to pinpoint in vivo the exact mechanisms that trigger ER stress. We previously hypothesized that several metabolic derangements in peroxisome-deficient Pex2−/− liver are likely to trigger ER stress, including perturbed flux of mevalonate metabolites, altered bile acid (BA) homeostasis, changes in fatty acid levels and composition, and oxidative stress [15]. Studies using genetic or dietary models of insulin resistance and fatty liver have demonstrated a key interconnectedness between hepatic steatosis and ER stress, as well as the physiological role of the UPR sensors in lipid homeostasis [60]. UPR activation has been observed in fatty liver diseases, although it is unclear how accumulation of excess lipids may engage ER stress response pathways [25]. Alternatively, UPR activation could occur before the onset of steatosis, and in fact steatosis may be a consequence of the UPR. While hepatic lipid accumulation could contribute to ER stress in postnatal SW/129 Pex2−/− mice, this is not the case in newborn Pex2−/− mice. Hepatic neutral lipid content is not increased in newborn mutants (Fig. 7), and protein levels of the lipid droplet protein ADFP were similar in P0 129 control and Pex2−/− livers (Fig. 7C).
Dysregulation of BA homeostasis has been linked to ER stress and UPR activation. Toxic hydrophobic bile acids are retained in the liver in cholestasis. In a genetic model of intrahepatic cholestasis, the accumulation of bile acids in the liver was associated with ER stress due to cumulative defects in expression of bile acid-CoA ligase, involved in BA conjugation, and BA transporters [61]. Cholestatic bile acid deposits were already observed in newborn Pex2−/− livers [13] and bile acid measurements in liver and plasma revealed an accumulation of mainly unconjugated C27 BAs and a deficiency of C24 BAs [13]. Hence, BA alterations in Pex2−/− mice could contribute to the activation of the UPR in the liver.
Alterations in cellular fatty acid composition may activate the UPR due to a disturbed physical state of cellular membranes and altered function and/or localization of membrane transport proteins. It has been shown that changes in fatty acid composition in stearoyl-CoA desaturase-1-deficient mice induce ER stress [62]. Long-chain free fatty acids can activate the UPR in several cell types [63–65]. Peroxisome deficiency leads to an accumulation of fatty acids that are degraded via peroxisomal β-oxidation (e.g., very long-chain and branched-chain fatty acids, dicarboxylic acids). Hepatic VLCFAs as well as some n-6 polyunsaturated fatty acids (docasahexaenoic acid, C22:6n-3) were significantly increased in newborn and P9 SW/129 Pex2−/− mice [10, 15].
Recent studies suggest that a subset of peroxisomal membrane proteins are inserted into the ER during the biogenesis of peroxisomes (class II peroxisomal membrane proteins) [66, 67], and retention of these proteins in the ER due to disturbed peroxisome assembly in Pex2−/− mice could potentially activate the UPR. However, we found that protein levels of several peroxins are reduced in the liver of both newborn and postnatal Pex2−/− mice (Fig. 5), and Pex14p was only found associated with peroxisomal membrane ghosts, which are characteristically larger in size, but fewer in number than normal peroxisomes [10, 68] (Fig. 6). Therefore, our studies suggest that peroxins are not retained in the ER and do not contribute to induction of ER stress in Pex2−/− liver.
Our studies further demonstrate an inverse relationship between induction of PPARα-regulated genes and SREBP-2-regulated genes in postnatal mice treated with the PPARα activator WY-14,643 (Fig. 4); this is also seen in global gene expression profiles in mouse liver in fasting-to-feeding and feeding-to-fasting transitions (W.J. Kovacs, unpublished results). Interestingly, hepatic FGF21 expression was significantly decreased by ~70% in newborn 129 and SW/129 Pex2−/− mice compared to controls, but is markedly induced in postnatal Pex2−/− liver (data not shown). In adult mice, FGF21 is induced directly by PPARα in liver in response to fasting and PPARα agonists [31–33], whereas hepatic FGF21 expression is very low in the fed state and in PPARα-deficient mice. The starvation faced by 129 Pex2−/− neonates is also accompanied by decreased FGF21 expression; however, the regulation of FGF21 in newborn or early postnatal mice has yet to be explored.
In summary, we have demonstrated that the SREBP-2 pathway and markers of ER stress and UPR activation, especially the integrated stress response branch, are markedly increased in the liver of newborn peroxisome-deficient 129 Pex2−/− mice. The ER stress in these newborn Pex2 mutants occurs in the absence of hepatic steatosis, peroxin accumulation or PPARα pathway induction. Given that prolonged or severe ER stress contributes to the pathogenesis of a number of human diseases, including diabetes, Alzheimer’s disease, and Parkinson’s disease, our findings suggest that functional peroxisomes are necessary to prevent chronic ER stress and dysregulation of the endogenous sterol response pathway.
Supplementary Material
Highlights.
Hepatic ER stress pathways are induced in peroxisome-deficient Pex2 knock-out mice.
ER stress deregulates the endogenous sterol response mechanism.
ER stress is induced in absence of hepatic steatosis or accumulation of peroxins.
The induction of SREBP-2 and ER stress is independent of PPARα activation.
Acknowledgments
This work was supported by NIH grants DK58238 and DK58040 to S.K.K., by NIH grants HD36807 and NS050602 to P.L.F., by NIH grant EY007361 to S.J.F., and by an Unrestricted Grant from Research to Prevent Blindness (S.J.F.). S.J.F. is the recipient of a Senior Scientific Investigator Award from Research to Prevent Blindness. K.N.C. was supported by a fellowship from SDSU-MARC funded by the National Institutes of General Medical Sciences/National Institutes of Health under grant T34GM008303 and by R01 minority supplement R01DK058238-03S1 to S.K.K. The work was also supported in part by the Swiss National Science Foundation (SNF) grant 31003A_132982 to W.J.K.
Abbreviations
- VLCFA
very long-chain fatty acid
- SREBP
sterol regulatory element-binding protein
- SCAP
SREBP cleavage-activating protein
- HMGCR
3-hydroxy-3-methylglutaryl-CoA reductase
- CoA
coenzyme A
- ER
endoplasmic reticulum
- Pex
peroxin
- HDL
high-density lipoprotein
- ISR
integrated stress response
- PERK
protein kinase RNA-like endoplasmic reticulum kinase
- ATF
activating transcription factor
- PPAR
peroxisome proliferator-activated receptor
- FPPS
farnesylpyrophosphate synthase
- IDI
isopentenyldiphosphate isomerase
- SQS
squalene synthase
- PFA
paraformaldehyde
- PBS
phosphate buffered saline
- RT
reverse transcription
- LDL
low-density lipoprotein
- SQLE
squalene epoxidase
- LSS
lanosterol synthase
- DHCR7
7-dehydrocholesterol reductase
- LDLR
LDL receptor
- Insig
insulin-induced gene
- CYP7A1
cholesterol 7α-hydroxylase
- CYP27A1
sterol 27-hydroxylase
- Grp
glucose-regulated protein
- GADD
growth arrest and DNA damage-inducible
- Herpud1
homocysteine-inducible ER stress-inducible ubiquitin-like domain member 1
- IRE1α
inositol-requiring enzyme-1
- XBP-1
X-box binding protein 1
- eIF2α
eukaryotic translation initiation factor 2
- CHOP
C/EBP homologous protein
- TRIB3
tribbles homolog 3
- ACOX1
acyl-CoA oxidase 1
- MFP1
multifunctional protein 1
- ADFP
adipose differentiation related protein
- CPT-1a
carnitine palmitoyltransferase 1a
- HMGCS
HMG-CoA synthase
- FGF21
fibroblast growth factor 21
- UPR
unfolded protein response
- TAG
triacylglycerol
- AMPK
AMP-activated protein kinase
- BA
bile acid
- UPR
unfolded protein response
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 2.Kovacs WJ, Olivier LM, Krisans SK. Central role of peroxisomes in isoprenoid biosynthesis. Prog Lipid Res. 2002;41:369–391. doi: 10.1016/s0163-7827(02)00002-4. [DOI] [PubMed] [Google Scholar]
- 3.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Yeagle PL. The Biology of Cholesterol. CRC Press; Boca Raton, FL: 1988. [Google Scholar]
- 6.Fliesler SJ. Sterols and Oxysterols: Chemistry, Biology and Pathobiology. Research Signpost; Kerala, India: 2002. [Google Scholar]
- 7.Kovacs WJ, Tape KN, Shackelford JE, Duan X, Kasumov T, Kelleher JK, Brunengraber H, Krisans SK. Localization of the pre-squalene segment of the isoprenoid biosynthetic pathway in mammalian peroxisomes. Histochem Cell Biol. 2007;127:273–290. doi: 10.1007/s00418-006-0254-6. [DOI] [PubMed] [Google Scholar]
- 8.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008;8:512–521. doi: 10.1016/j.cmet.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faust PL, Hatten ME. Targeted deletion of Pex2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J Cell Biol. 1997;139:1293–1305. doi: 10.1083/jcb.139.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faust PL, Su HM, Moser A, Moser HW. The peroxisome deficient Pex2 Zellweger mouse. J Mol Neurosci. 2001;16:289–297. doi: 10.1385/JMN:16:2-3:289. [DOI] [PubMed] [Google Scholar]
- 12.Faust PL. Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J Comp Neurol. 2003;461:394–413. doi: 10.1002/cne.10699. [DOI] [PubMed] [Google Scholar]
- 13.Keane MH, Overmars H, Wikander TM, Ferdinandusse S, Duran M, Wanders RJA, Faust PL. Bile acid treatment alters hepatic disease and bile acid transport in peroxisome-deficient Pex2 Zellweger mice. Hepatology. 2007;45:982–997. doi: 10.1002/hep.21532. [DOI] [PubMed] [Google Scholar]
- 14.Kovacs WJ, Shackelford JE, Tape KN, Richards MJ, Faust PL, Fliesler SJ, Krisans SK. Disturbed cholesterol homeostasis in a peroxisome-deficient Pex2 knockout mouse model. Mol Cell Biol. 2004;24:1–13. doi: 10.1128/MCB.24.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacs WJ, Tape KN, Shackelford JE, Wikander TM, Richards MJ, Fliesler SJ, Krisans SK, Faust PL. Peroxisome deficiency causes a complex phenotype because of hepatic SREBP/Insig dysregulation associated with endoplasmic reticulum stress. J Biol Chem. 2009;284:7232–7245. doi: 10.1074/jbc.M809064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leuenberger N, Pradervand S, Wahli W. Sumoylated PPARa mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J Clin Invest. 2009;119:3138–3148. doi: 10.1172/JCI39019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.König B, Koch A, Spielmann J, Hilgenfeld C, Stangl GI, Eder K. Activation of PPARa lowers synthesis and concentration of cholesterol by reduction of nuclear SREBP-2. Biochem Pharmacol. 2007;73:574–585. doi: 10.1016/j.bcp.2006.10.027. [DOI] [PubMed] [Google Scholar]
- 18.Knight BL, Hebbachi A, Hauton D, Brown AM, Wiggins D, Patel DD, Gibbons GF. A role for PPARa in the control of SREBP activity and lipid synthesis in the liver. Biochem J. 2005;389:413–421. doi: 10.1042/BJ20041896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiota Y, Ikeda M, Hashimoto F, Hayashi H. Effects of peroxisome proliferators gemfibrozil and clofibrate on syntheses of dolichol and cholesterol in rat liver. J Biochem (Tokyo) 2003;134:197–202. doi: 10.1093/jb/mvg129. [DOI] [PubMed] [Google Scholar]
- 20.Roglans N, Peris C, Verd JC, Alegret M, Vazquez M, Sanchez RM, Laguna JC. Increase in hepatic expression of SREBP-2 by gemfibrozil administration to rats. Biochem Pharmacol. 2001;62:803–809. doi: 10.1016/s0006-2952(01)00701-8. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto F, Taira S, Hayashi H. Changes in isoprenoid lipid synthesis by gemfibrozil and clofibrate in rat hepatocytes. Biochem Pharmacol. 2000;59:1203–1210. doi: 10.1016/s0006-2952(00)00261-6. [DOI] [PubMed] [Google Scholar]
- 22.Dirkx R, Vanhorebeek I, Martens K, Schad A, Grabenbauer M, Fahimi D, Declercq P, van Veldhoven PP, Baes M. Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology. 2005;41:868–878. doi: 10.1002/hep.20628. [DOI] [PubMed] [Google Scholar]
- 23.Anderson SP, Dunn C, Laughter A, Yoon L, Swanson C, Stulnig TM, Steffensen KR, Chandraratna RAS, Gustafsson JA, Corton JC. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor α, retinoid X receptor, and Liver X receptor in mouse liver. Mol Pharmacol. 2004;66:1440–1452. doi: 10.1124/mol.104.005496. [DOI] [PubMed] [Google Scholar]
- 24.Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Müller M, Kersten S. Comprehensive analysis of PPARα-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee AH, Glimcher LH. Intersection of the unfolded protein response and hepatic lipid metabolism. Cell Mol Life Sci. 2009;66:2835–2850. doi: 10.1007/s00018-009-0049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheehan DC, Hrapchak BB. Theory and practice of histotechnology. Battelle Press; Columbus, OH: 1980. [Google Scholar]
- 27.Burg JS, Espenshade PJ. Regulation of HMG-CoA reductase in mammals and yeast. Prog Lipid Res. 2011;50:403–410. doi: 10.1016/j.plipres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martens K, van Themaat EVL, van Batenburg MF, Heinäniemi M, Huyghe S, van Hummelen P, Carlberg C, van Veldhoven PP, van Kampen A, Baes M. Coordinate induction of PPARα and SREBP2 in multifunctional protein 2 deficient mice. Biochim Biophys Acta. 2008;1781:694–702. doi: 10.1016/j.bbalip.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Dalen KT, Ulven SM, Arntsen BM, Solaas K, Nebb HI. PPARα activators and fasting induce the expression of adipose differentiation-related protein in liver. J Lipid Res. 2006;47:931–943. doi: 10.1194/jlr.M500459-JLR200. [DOI] [PubMed] [Google Scholar]
- 30.Edvardsson U, Ljungberg A, Linden D, William-Olsson L, Peilot-Sjogren H, Ahnmark A, Oscarsson J. PPARalpha activation increases triglyceride mass and adipose differentiation-related protein in hepatocytes. J Lipid Res. 2006;47:329–340. doi: 10.1194/jlr.M500203-JLR200. [DOI] [PubMed] [Google Scholar]
- 31.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, Elmquist JK, Gerard RD, Burgess SC, Hammer RE, Mangelsdorf DJ, Kliewer SA. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Lundasen T, Hunt MC, Nilsson LM, Sanyal S, Angelin B, Alexson SE, Rudling M. PPARalpha is a key regulator of hepatic FGF21. Biochem Biophys Res Commun. 2007;360:437–440. doi: 10.1016/j.bbrc.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 34.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harding HP, Zeng H, Zhang Y, Jungreis R, Chung P, Plesken H, Sabatini DD, Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 36.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 37.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 38.Lu PD, Harding HP, Ron D. Translation re-initiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passe CM, Cooper G, Quirk CC. The murine p8 gene promoter is activated by activating transcription factor 4 (ATF4) in the gonadotrope-derived LbetaT2 cell line. Endocrine. 2006;30:81–91. doi: 10.1385/endo:30:1:81. [DOI] [PubMed] [Google Scholar]
- 42.Titorenko VI, Rachubinski RA. Spatiotemporal dynamics of the ER-derived peroxisomal endomembrane system. Int Rev Cell Mol Biol. 2009;272:191–244. doi: 10.1016/S1937-6448(08)01605-5. [DOI] [PubMed] [Google Scholar]
- 43.Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolins NE, Brasaemle DL, Bickel PE. A proposed model of fat packaging by exchangeable lipid droplet proteins. FEBS Lett. 2006;580:5484–5491. doi: 10.1016/j.febslet.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 46.Xu G, Sztalryd C, Lu X, Tansey JT, Gan J, Dorward H, Kimmel AR, Londos C. Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J Biol Chem. 2005;280:42841–42847. doi: 10.1074/jbc.M506569200. [DOI] [PubMed] [Google Scholar]
- 47.Xu G, Sztalryd C, Londos C. Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim Biophys Acta. 2006;1761:83–90. doi: 10.1016/j.bbalip.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 48.Vanhorebeek I, Baes M, Declercq PE. Isoprenoid biosynthesis is not compromised in a Zellweger syndrome mouse model. Biochim Biophys Acta. 2001;1532:28–36. doi: 10.1016/s1388-1981(01)00108-1. [DOI] [PubMed] [Google Scholar]
- 49.Hogenboom S, Romeijn GJ, Houten SM, Baes M, Wanders RJA, Waterham HR. Absence of functional peroxisomes does not lead to deficiency of enzymes involved in cholesterol biosynthesis. J Lipid Res. 2002;43:90–98. [PubMed] [Google Scholar]
- 50.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, Maeda N, Krisans SK, Malinow MR, Austin RC. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 52.Colgan SM, Tang D, Werstuck GH, Austin RC. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 53.Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal. 2010;8:e002. doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hostetler HA, Petrescu AD, Kier AB, Schroeder F. Peroxisome proliferator-activated receptor α interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–18682. doi: 10.1074/jbc.M412062200. [DOI] [PubMed] [Google Scholar]
- 55.Cook WS, Jain S, Jia Y, Cao WQ, Yeldandi AV, Reddy JK, Rao MS. Peroxisome proliferator-activated receptor α-responsive genes induced in the newborn but not prenatal liver of peroxisomal fatty acyl-CoA oxidase null mice. Exp Cell Res. 2001;268:70–76. doi: 10.1006/excr.2001.5266. [DOI] [PubMed] [Google Scholar]
- 56.Wu J, Rutkowski DT, Dubois M, Swathirajan J, Saunders T, Wang J, Song B, Yau GD, Kaufman RJ. ATF6a optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 58.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 59.Huang J, Viswakarma N, Yu S, Jia Y, Bai L, Vluggens A, Cherkaoui-Malki M, Khan M, Singh I, Yang G, Rao MS, Borensztajn J, Reddy JK. Progressive endoplasmic reticulum stress contributes to hepatocarcinogenesis in fatty acyl-CoA oxidase 1-deficient mice. Am J Pathol. 2011;179:703–713. doi: 10.1016/j.ajpath.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bochkis IM, Rubins NE, White P, Furth EE, Friedman JR, Kaestner KH. Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress. Nat Med. 2008;14:828–836. doi: 10.1038/nm.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Flowers MT, Keller MP, Choi Y, Lan H, Kendziorski C, Ntambi JM, Attie AD. Liver gene expression analysis reveals endoplasmic reticulum stress and metabolic dysfunction in SCD1-deficient mice fed a very low-fat diet. Physiol Genomics. 2008;33:361–372. doi: 10.1152/physiolgenomics.00139.2007. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y, Wang D, Gentile CL, Pagliassotti MJ. Reduced endoplasmic reticulum luminal calcium links saturated fatty acid-mediated endoplasmic reticulum stress and cell death in liver cells. Mol Cell Biochem. 2009;331:31–40. doi: 10.1007/s11010-009-0142-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pfaffenbach KT, Gentile CL, Nivala AM, Wang D, Wei Y, Pagliassotti MJ. Linking endoplasmic reticulum stress to cell death in hepatocytes: roles of C/EBP homologous protein and chemical chaperones in palmitate-mediated cell death. Am J Physiol Endocrinol Metab. 2010;298:E1027–E1035. doi: 10.1152/ajpendo.00642.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cazanave SC, Elmi NA, Akazawa Y, Bronk SF, Mott JL, Gores GJ. CHOP and AP-1 cooperatively mediate PUMA expression during lipoapoptosis. Am J Physiol Gastrointest Liver Physiol. 2010;299:G236–G243. doi: 10.1152/ajpgi.00091.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Platta HW, Erdmann R. Peroxisomal dynamics. Trends Cell Biol. 2007;17:474–484. doi: 10.1016/j.tcb.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 67.Ma C, Agrawal G, Subramani S. Peroxisome assembly: matrix and membrane protein biogenesis. J Cell Biol. 2011;193:7–16. doi: 10.1083/jcb.201010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santos MJ, Imanaka T, Shio H, Small GM, Lazarow PB. Peroxisomal membrane ghosts in Zellweger syndrome – aberrant organelle assembly. Science. 1988;239:1536–1538. doi: 10.1126/science.3281254. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.