Abstract
Chinese herbal medicinal (CHM) extracts from fourteen plants were investigated in cell-based in vitro assays for their effect on nuclear factor κB (NF-κB), a key regulator of inflammation, as well as on peroxisome proliferator-activated receptors (PPARs) being key regulators of genes involved in lipid and glucose metabolism. 43% of the investigated CHMs showed NF-κB inhibitory and 50% PPARα and PPARγ activating effects. Apolar extracts from cortex and flos of Albizia julibrissin Durazz. and processed rhizomes of Arisaema sp. and Pinellia ternata (Thunb.) Breit. that effectively inhibited TNF-α-induced NF-κB activation and dose-dependently activated PPARα and PPARγ were further investigated. Bioassay-guided fractionation and analysis by GC-MS led to the identification of fatty acids as PPAR agonists, including linoleic and palmitic acid.
1. Introduction
Herbal medicines are an important part of Traditional Chinese Medicine (TCM) of which the medical use and processing methods are well documented. Traditional processing methods (pao zhi) are important to enhance the efficacy and/or to reduce the toxicity of crude herbal products [1]. Chinese herbal medicine (CHM) encompasses over 11,000 species of medicinal plants and is a valuable source [2] for the identification of biologically active natural products. Investigation of the molecular targets and mechanistic action of CHMs and their single compounds is currently a central task in TCM research [3]. Thanks to advances in molecular biology, refined bioassays are now available which enable rapid screening of natural products for bioactivity towards specific targets [4]. In this study, extracts of different polarity from CHMs of fourteen plant species were tested for a potential inhibition of TNF-α-induced NF-κB activation and an agonistic activity towards PPARα and PPARγ. The CHMs were selected in cooperation with Chinese partners [5]. This in-house collection of CHMs (Table 1) was already examined earlier based on their traditional use against insomnia and anxiety regarding a putative modulation of the GABAA receptor [6]. The listed CHMs are, however, also traditionally prescribed as single herbs as well as in formulations for clearing heat and drying dampness, among others. Therefore, to gain insight into possible multi-target effects of the individual CHMs, this study examined their influence on the nuclear factor κB (NF-κB) pathway and peroxisome proliferator-activated receptors (PPARs), which are important drug targets with regard to inflammation and metabolic dysfunction.
Table 1.
The plant species and the respective Chinese herbal medicines tested for potential activation of human PPARα and PPARγ and inhibition of TNF-α-induced NF-κB activity.
| Plant species | Family | Plant part | Pin Yin | CHM no. |
|---|---|---|---|---|
| Albizia julibrissin Durazz. | Fabaceae | Cortex | hé huān pí | 1a |
| Flos | hé huān huā | 1b | ||
| Arisaema sp. | Araceae | Rhizoma* | zhì tiān nán xīng | 2a |
| Rhizoma** | zhì tiān nán xīng | 2b | ||
| Arnebia/Lithospermum sp. | Boraginaceae | Radix | zǐ cǎo | 3 |
| Atractylodes macrocephala Koidz. | Asteraceae | Rhizoma | bái zhú | 4 |
| Cnidium monnieri (L.) Cuss. | Apiaceae | Fructus | shé chuáng zǐ | 5 |
| Dimocarpus (Euphoria) longan Lour. | Sapindaceae | Arillus | long yǎn ròu | 6 |
| Forsythia suspensa (Thunb.) Vahl. | Oleaceae | Fructus | lián qiào | 7 |
| Juncus effusus L. | Juncaceae | Medulla | dēng xīn cǎo | 8 |
| Lilium sp. | Liliaceae | Bulbus | bǎi hé | 9 |
| Lophatherum gracile Brongn. | Poaceae | Herba | dàn zhú yè | 10 |
| Nelumbo nucifera Gaertn. | Nelumbonaceae | Plumula | lián zǐ xīn | 11 |
| Pinellia ternata (Thunb.) Breit. | Araceae | Rhizoma | bàn xià | 12a |
| Rhizoma** | zhì bàn xià | 12b | ||
| Polygonum multiflorum Thunb. | Polygonaceae | Caulis | shŏu wū téng | 13 |
| Tribulus terrestris L. | Zygophyllaceae | Fructus | cì jí lí | 14 |
Processed by cooking * or soaking ** in water with alum (KAl(SO4)2·12H2O).
The NF-κB signaling pathway is a key regulator of inflammation. Inflammatory stimuli such as tumor necrosis factor alpha (TNF-α), infectious agents (lipopolysaccharide (LPS)), injury, and other stressful conditions activate the NF-κB signal transduction pathway. Thereby, activation of the IKK complex leads to phosphorylation and subsequent proteasomal degradation of IκB proteins. NF-κB dimers are translocated to the nucleus and bind to κB promotor sites resulting in the transcription of proinflammatory target genes [7, 8]. Inhibitors of NF-κB signaling are promising candidates for both the prevention and therapy of (chronic) inflammation. Terpenoids, such as parthenolide, are known to be effective natural NF-κB inhibitors [9].
PPARs regulate the expression of genes involved in lipid and glucose metabolism and homeostasis in a ligand-dependent manner [10]. In the activated form, PPARα promotes mainly fatty acid (FA) catabolism, and PPARγ enhances insulin sensitivity and lipid storage [11, 12]. PPARα and PPARγ agonism is also linked to the suppression of pro-inflammatory genes, for example, via an interference with the NF-κB signaling pathway [13, 14]. Synthetic PPAR agonists (fibrates and thiazolidinediones) bind PPARs with high affinity. They, however, show significant dose limiting adverse side effects. Natural products were revealed to be a promising source of safer PPARα and PPARγ dual agonists or partial PPAR modulators [15–18].
In our study, extracts of selected CHMs that exhibited a significant activity towards PPARs or NF-κB were further subjected to dose-response studies and bioassay-guided fractionation. These selected CHMs were cortex and flos of Albizia julibrissin Durazz. as well as processed and crude rhizomes of Arisaema sp. and Pinellia ternata (Thunb.) Breit. Traditionally, A. julibrissin cortex and flos are used in CHM to treat insomnia and injuries and to calm the mind. The rhizomes of Arisaema sp. and P. ternata are toxic in their crude form and therefore traditionally processed (Table 1). Processed rhizomes of Arisaema sp. and P. ternata CHMs are effective in removing damp-phlegm such as in convulsions and spasms in the intestines [19, 20]. The dried, pulverized crude rhizomes from P. ternata are used externally to treat swellings. Bioactive compounds were isolated, analyzed, and characterized by mass spectrometry and NMR.
2. Materials and Methods
2.1. Materials
Human embryonic kidney (HEK) 293 cells stably transfected with an NF-κB-responsive luciferase reporter were purchased from Panomics (Fremont, CA, USA) and 3T3-L1 preadipocytes and HEK293 cells from the American Type Culture Collection (Manassas, VA, USA). The PPAR luciferase reporter construct (tk-PPREx3-luc) and the expression plasmids for PPARα and PPARγ (pCMX-mPPARα and pCMX-mPPARγ) were a gift from Professor Ronald M. Evans (Howard Hughes Medical Institute, La Jolla, CA, USA). The plasmid encoding enhanced green fluorescent proteins (pEGFP-N1) was obtained from Clontech (Mountain View, CA, USA). Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/L glucose was purchased from Lonza Group AG (Basel, Switzerland) and fetal bovine serum (FBS) from Invitrogen (Lofer, Austria). GW7647 and troglitazone were purchased from Cayman Europe (Tallinn, Estonia) and 2-deoxy-D-(1H3)-glucose from Perkin Elmer (Waltham, MA, USA). For extraction, fractionation, and isolation by silica column chromatography (CC), solvents of highest available purity were used (VWR, Vienna, Austria). All other chemicals were obtained from Sigma-Aldrich (Vienna, Austria).
2.2. Plant Materials
Material of the CHMs no. 1–14 were purchased from Plantasia (Oberndorf, Austria) (see Table 1) [19, 20], except of no. 12a which was purchased from Sichuan Neautus Traditional Chinese Medicine Co., Ltd (Chengdu, China). Voucher specimens are deposited at the Department of Pharmacognosy, University of Vienna, encoded with the CHM numbers shown in Table 1.
2.3. Extraction and Fractionation
50 g of each of the CHMs nos. 1a, 1b, 2a, 3–11, 12b, 13, and 14 was soaked in 500 mL petroleum ether (PE) for 10 min, and extracted under reflux for 30 min. The obtained extracts were filtered and evaporated to dryness. The remaining drug material was air-dried overnight and extracted each with 500 mL of ethyl acetate (EtOAc), methanol (MeOH), and distilled water likewise, whereas the water extract was lyophilized. All following extractions were performed at 150 bar and 40°C using an ASE 200 accelerated solvent extractor and a solvent controller (Dionex, Sunnyvale, CA, USA). With the ASE every ca. 10 g of pulverized plant material in a capsule was extracted three times with 22 mL of solvent (66 mL/ca. 10 g). The scheme of each run of extraction was as follows: 5 min heating, 2 min static at 150 bar and 40°C, 10 s flushing and 60 s purging. For CHMs nos. 2b, 12a, and 12b out of 50 g each DCM dry extract yields of 0.4–0.7% were obtained and for no. 1b a yield of 2.9% (See Schemes 1 and 2 in Supplementary Material available online at doi:10.1155/2012/983023).
For bioassay-guided fractionation, 1.4 kg of CHMs no. 1a and 1.9 kg of no. 2a were pulverized and extracted by DCM and subsequently by MeOH. 68 g DCM extract (4.7% yield) and 92 g MeOH extract (6.6% yield) were obtained of no. 1a. 12.1 g DCM (0.6% yield) and 10.7 g MeOH (0.8% yield), out of 1.4 kg of the material already extracted by DCM, were obtained of no. 2a. The dried extracts of no. 2a were subjected to silica CC with Silica Gel 60 (particle size of 0.063–0.200 mm, Merck, Darmstadt, Germany). The column with DCM extract was eluted with CHCl3 : MeOH : H2O 98 : 2 : 1 to 60 : 38 : 8.5 to obtain eight fractions (D1–D8). From fractions D8, cerebrosides were isolated by silica CC eluting with CHCl3 : MeOH : H2O 65 : 25 : 4. Cerebrosides were also detected in CHMs no. 12a and b from P. ternata. 15 g of DCM extract of no. 1a was fractionated using VLC (Vacuum Liquid Chromatography) using Silica Gel 60 (particle size of 0.063–0.200 mm, Merck, Darmstadt, Germany) as solid phase. The silica column was eluted subsequently with PE, DCM (twice), EtOAc, MeOH, and 90% MeOH (3 L per solvent) under reduced pressure. Thereby 12 mg PE extract, 963 mg extract from the first 3L eluted with DCM (DCM-I), 776 mg from the next 3L eluted with DCM (DCM-II), 6.90 g EtOAc extract, 4.08 g MeOH extract, and finally 170 mg extract eluted with 90% MeOH were obtained as dry extracts. Betulinic acid (12 mg) was isolated from EtOAc VLC fraction of the DCM extract from no. 1a. A mixture of trihydroxy and epoxy-dihydroxy fatty acid methyl esters (FAMEs) (8 mg) and α-spinasterol-3-O-β-D-glucopyranoside (61 mg) was isolated from the crude MeOH extract by repeated silica CC eluting with CHCl3 : MeOH : H2O and n-hexane : EtOAc gradients. The compounds in the mixture could be separated by GC-MS and were identified based on resemblance with the fragmentation in EI MS reported by Hamberg [21] as methyl 9,10,13-trihydroxy-11-octadecenoate, methyl 9,12,13-trihydroxy-11-octadecenoate, and methyl 9,10-epoxy-13-hydroxy-11-octadecenoate. Their hydroxylated FA derivatives were prepared by basic hydrolysis with 0.1 M NaOH (15 min RT) of the hydroxylated FAMEs and acidification with 0.1 M HCl/MeOH. For biological testing from all corresponding samples, always dry material was dissolved in DMSO.
2.4. NMR Analysis
All 1D (1H and 13C) and 2D (COSY, NOESY, HMQC, and HMBC) NMR spectra were recorded on a Bruker Avance 500 NMR spectrometer. Betulinic acid was dissolved in CDCl3 (99.96 atom% D) and α-spinasterol-3-O-β-D-glucopyranoside in d5-pyridine (99.96 atom% D). The 1H and 13C NMR spectra were operated at 500 and 125 MHz, respectively. 13C and 1H NMR data of both were consistent with the literature values for betulinic acid [22, 23] and α-spinasterol-3-O-β-D-glucopyranoside [24, 25].
2.5. Analysis of Active Compounds by GC-MS
The investigated extracts and fractions were dissolved to 5 mg/mL in DCM, and 200 μL of the dissolved sample was transferred to a GC-MS vial, to which 50 μL of TMSH was added. The mixture with a concentration of 4 mg/mL was shaken for 30 s. A GCMS-QP2010 gas chromatograph mass spectrometer was utilized (Shimadzu Scientific Instruments, Columbia, MD, USA). Helium 5.0 was used as carrier gas, and the column flow was set at a constant value of 1.7 mL/min. The column used was a Zebron ZB-5 60 m × 0.25 mm (inner diameter), 0.25 μm (film thickness) (Phenomenex, Torrance, CA, USA). The software used was Lab Solutions GC-MS (version 2.50 SU3, Shimadzu). 1 μL of sample was injected of the prepared solutions with a split 1 : 10, and analysis was performed in the EI mode (70 eV, 250°C ion source temperature). The injection temperature was 270°C. A method with a temperature gradient from 120°C to 320°C in 40 min and 5 min hold on 320°C was applied for analysis. A mixture of trihydroxy- and epoxy-hydroxy FAMEs from no. 1a was derivatised by BSTFA-TMCS (99 : 1) for 30 min at 50°C and detected by GC-MS in the same conditions as above. The analyses were performed on an Agilent Technologies 6890 N Network GC equipped with an Agilent Technologies 5973 inert mass selective detector and a Combi PAL autosampler (CTC Analytics). The column used was a DB-5 with dimensions of 30 m × 0.25 mm (inner diameter), 0.23 μm (film thickness) (Agilent Technologies). The software used was MSD Chemstation 2004.
2.6. NF-κB Transactivation Assay
HEK 293 cells, stably transfected with as NF-κB-responsive luciferase reporter (Panomics, Fremont, CA, USA), were seeded in 10 cm dishes and transfected with 5 μg pEGFP-N1 (Clontech, Mountain View, CA, USA). Six hours later, cells were transferred to 96-well plates and incubated overnight (5% CO2, 38°C). On the next day, the medium was exchanged with a serum-free DMEM, and cells were treated with the indicated extracts or fractions. The solvent vehicle (0.1% DMSO) served as negative control and 5 μM parthenolide as positive control. After one hour, cells were stimulated with 2 ng/mL human recombinant TNF-α for 6 h, then the medium was removed, and cells were lysed. The luminescence of the firefly luciferase and the fluorescence of the enhanced green fluorescent protein (EGFP) were quantified on a GeniosPro plate reader (Tecan, Austria). The luciferase signal derived from the NF-κB reporter was normalized with the EGFP-derived fluorescence to account for differences in the cell number.
2.7. PPAR Luciferase Reporter Gene Assay
The PPAR-luciferase reporter gene assay was performed as previously described [26]. HEK293 cells were transiently transfected with a PPARα or PPARγ receptor expression plasmid, a reporter plasmid (tk-PPREx3-luc), and a green fluorescent protein plasmid (pEGFP-N1) as an internal control. The cells were harvested 6 h after the transfection and reseeded in 96-well plates (5 × 104 cells/well). In the initial tests, cells were treated with 10 μg/mL of the indicated extracts. In the dose-response experiments, cells were treated with 0, 3, 27 or 81 μg/mL of the DCM extracts of CHMs nos. 1a, 1b, 2a, 2b, 12a, and 12b. To account for potential effects of the solvent, 0.1% DMSO served as vehicle control. As positive controls, 50 nM GW7647 and 5 μM troglitazone were used to activate PPARα and PPARγ, respectively. Treated cells were incubated for 18 h. After cell lysis, the luminescence of the firefly luciferase and the fluorescence of EGFP were quantified on a GeniosPro plate reader (Tecan, Salzburg, Austria). The luminescence signals were normalized to the EGFP-derived fluorescence to account for differences in cell number or transfection efficiency.
2.8. Statistical Analysis
Statistical analysis was performed using Prism Software (ver. 4.03; GraphPad Software Inc., San Diego, CA). Data were normalized to DMSO-treated control of which the mean value was set as 1.0. The data shown represent arithmetic mean and standard deviation of 3-4 independent experiments. Statistical significance was determined by a one-way analysis of variance combined with a Dunnett's Multiple Comparison posttest. Results with P < 0.05 were considered significant.
3. Results and Discussion
In this study, extracts of important Chinese herbal medicines from fourteen different plant species (Table 1) were tested for their potential to inhibit NF-κB and/or to activate PPARα and PPARγ.
Extracts of CHMs of six out of fourteen plant species reduced TNF-α-induced NF-κB activity significantly (P < 0.05) (Table 2). In fact, this is the first report of a significant NF-κB inhibitory activity of extracts from A. julibrissin, Arisaema sp., J. effuses, and P. ternata. A moderate (see explanation accompanying Table 2) reduction of TNF-α-induced NF-κB activity (P < 0.05) was found for the PE, EtOAc, and MeOH extracts of A. julibrissin no.1a and the EtOAc extract of no. 1b and C. monnieri no. 5, and PE extracts of Arisaema sp. no. 2a and P. ternata no. 12b. The EtOAc extracts of F. suspensa no. 7, J. effusus no. 8, and the aqueous extract of A. julibrissin no. 1b exhibited an especially strong ability (see explanation accompanying Table 2) to inhibit NF-κB activity. Betulinic acid was isolated from the DCM extract of the bark of A. julibrissin no. 1a [27]. Many terpenoids, including betulinic acid, are reported to effectively inhibit NF-κB signaling [9, 28]. However, we neither detected NF-κB inhibition nor PPARα and PPARγ activation by betulinic acid in concentrations up to 30 μM excluding betulinic acid as one of the main active principles in our cellular models. Osthole might in part have contributed to the observed activities of the EtOAc extract of C. monnieri no. 5 [29–31].
Table 2.
Screening of Chinese herbal medicines for agonistic activity towards PPARα and PPARγ and inhibition of TNF-α-induced NF-κB activity in vitro. PE = petroleum ether, EtOAc = ethyl acetate, MeOH = methanol. Dry material from all test samples was dissolved in DMSO at a concentration of 10 mg/mL and tested at a final concentration of 10 μg/mL. Regarding PPARs: no activation compared to control (0.1% DMSO): −; 1.5- to 2-fold activity compared to control (P < 0.05): +; more than 2-fold of the control (P < 0.05): ++. Regarding NF-κB: inhibition in the range of 0–50% compared to the vehicle-treated control (0.1% DMSO): −; 50–80% inhibition (P < 0.05): +; over 80% inhibition (P < 0.05): ++. At least three independent experiments were performed with every sample, and statistical analysis was performed by ANOVA.
| Plant species | CHM no. | Extract | PPARα | PPARγ | NF-κB |
|---|---|---|---|---|---|
| activation | activation | inhibition | |||
| Albizia julibrissin Durazz. | 1a | PE | ++ | ++ | + |
| EtOAc | ++ | ++ | + | ||
| MeOH | + | + | + | ||
| H2O | − | − | + | ||
| 1b | PE | + | + | − | |
| EtOAc | ++ | ++ | + | ||
| MeOH | − | − | − | ||
| H2O | − | − | ++ | ||
|
| |||||
| Arisaema sp. | 2a | PE | ++ | + | + |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Arnebia/Lithospermum sp. | 3 | PE | − | − | − |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Atractylodes macrocephala Koidz. | 4 | PE | − | − | − |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Cnidium monnieri (L.) Cuss. | 5 | PE | − | − | − |
| EtOAc | ++ | + | + | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Dimocarpus (Euphoria) longan Lour. | 6 | PE | − | − | − |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Forsythia suspensa (Thunb.) Vahl. | 7 | PE | − | − | + |
| EtOAc | − | − | ++ | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Juncus effusus L. | 8 | EtOAc | − | − | ++ |
| MeOH | − | − | + | ||
| H2O | − | − | − | ||
|
| |||||
| Lilium sp. | 9 | EtOAc | + | − | − |
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Lophatherum gracile Brongn. | 10 | PE | + | − | − |
| EtOAc | + | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Nelumbo nucifera Gaertn. | 11 | PE | − | − | − |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Pinellia ternata (Thunb.) Breit. | 12b | PE | ++ | + | + |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Polygonum multiflorum Thunb. | 13 | PE | − | − | − |
| EtOAc | − | − | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
|
| |||||
| Tribulus terrestris L. | 14 | PE | − | − | − |
| EtOAc | + | + | − | ||
| MeOH | − | − | − | ||
| H2O | − | − | − | ||
When testing for a PPARα and PPARγ agonistic activity, a significant effect (P < 0.05) was observed for apolar extracts of seven out of the fourteen plants used as CHM (Table 2). Moderate to strong PPARα and PPARγ activity was observed for apolar extracts from A. julibrissin no. 1a and 1b, Arisaema sp. no. 2a, C. monnieri no. 5, P. ternata no. 12a and T. terrestris no. 14 (P < 0.05). Apolar extracts of Lilium sp. no. 9 and L. lophatherum no. 10 activated PPARα moderately (P < 0.05). Previously, a saponin from fruits of T. terrestris [32, 33] and aqueous extracts of arillus from D. longan and rhizomes of P. ternata [34–36] were reported to increase PPARα and PPARγ gene expression levels.
A selection of CHMs with strong activity, namely, A. julibrissin no. 1a and b, Arisaema sp. no. 2a and b and of P. ternata no. 12a, and b were further studied. The DCM extracts of no. 1a, 1b, 2a, 2b, 12a, and 12b showed a significant dose-dependent increase of PPARα and PPARγ activity (P < 0.01) (Figure 1), whereby those of nos. 1a and 1b from A. julibrissin and no. 2a from Arisaema sp. were most potent. Loss of activity at 81 μg/mL of these extracts was possibly due to toxicity or unspecific inhibitory action.
Figure 1.
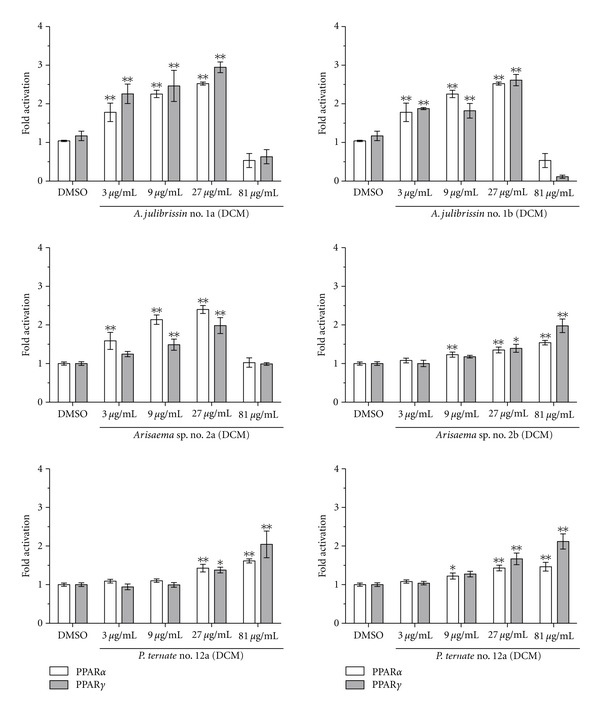
Dose-response experiments with DCM extracts of CHMs of A. julibrissin no. 1a and b, Arisaema sp. no. 2a and b, and P. ternata no. 12a and b. HEK293 cells were transiently transfected with the expression plasmid for PPARα or PPARγ, the reporter plasmid pPPRE-tk3x-luc, and the internal control plasmid (EGFP). The transiently transfected cells were incubated for 18 h with 3, 9, 27, and 81 μg/mL of each indicated extract. Furthermore, the cells were similarly incubated with 0.1% DMSO (negative control), 50 nM GW7647 (PPARα agonist, activated PPARα 2.5–3.5 fold but not PPARγ), or 5 μM troglitazone (PPARγ agonist, activated 3.5–6.5 fold PPARγ but not PPARα) as positive controls (not shown). Luciferase activity and fluorescence intensity were measured. Results are presented as mean ± SD, (n = 4). Significantly different from the negative control (ANOVA), *P < 0.05; **P < 0.01.
Bioassay-guided fractionation showed that the fraction “DCM-II” (see Section 2) of the DCM extract from A. julibrissin no. 1a significantly activated PPARα by 3.5-fold (±0.31, P < 0.01) and PPARγ by 2.8-fold (±0.28, P < 0.01) (compared to DMSO as solvent control). The EtOAc VLC fraction significantly activated PPARα by 2.9-fold (±0.52, P < 0.01) and PPARγ by 2.5-fold (±0.19, P < 0.01). The apolar fraction “D6” from Arisaema sp. no. 2a significantly activated PPARα by 2.2-fold (±0.10, P < 0.01) and PPARγ by 1.5-fold (±0.06, P < 0.01) (Figure 2). GC-MS analysis after derivatisation revealed that the VLC fractions of A. julibrissin no. 1a (DCM) and fraction “D6” from Arisaema sp. no. 2a contained mainly fatty acids (FAs), such as palmitic acid (16 : 0), stearic acid (18 : 0), and linoleic acid (18 : 2) (Figures 3 and 4) that are known as PPAR agonists [11]. 13-Phenyltridecanoic acid (13 : 0) was found as a compound specific for aroids [37]. Especially polyunsaturated FAs activate PPARs potently [38]. Wolfrum et al. reported palmitic acid (18 : 0), linoleic acid (18 : 2), oleic acid (18 : 1), and stearic acid (18 : 0) to activate PPARα by 2.5-, 4.6-, 3.5-, and 2-fold, respectively [39]. The processing might have influenced availability of FA from rhizomes of P. ternata. The concentration of FAs detected was higher in the DCM extract of processed rhizomes of P. ternata (no. 12b) than in that of the crude rhizomes (no. 12a) (Figure 4). This was in line with the observation that the DCM extract of the processed 12b was PPARα and PPARγ active at slightly lower concentration than that of the crude rhizomes (no. 12a) (Figure 1). The activation of PPARα by fraction “D6” may be attributed largely to palmitic acid, which is highly enriched in fraction “D6” reaching 61% of the total content (Figure 4). α-Spinasterol-3-O-β-D-glucopyranoside from A. julibrissin and cerebrosides isolated from the DCM extract of rhizomes of Arisaema sp. and P. ternata were tested; however, did not show agonistic activity towards PPARα- and PPARγ-activity in vitro.
Figure 2.
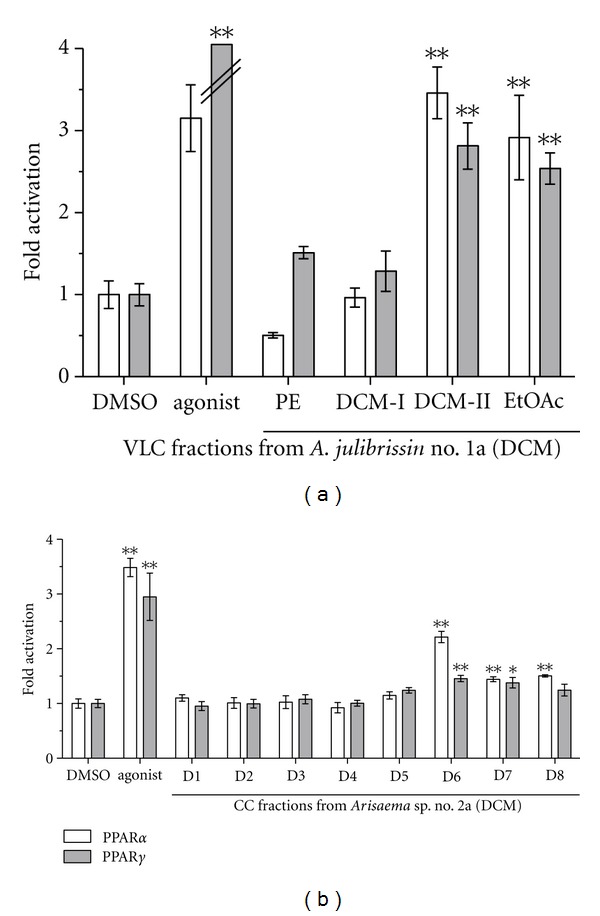
Comparison of the PPARα- and PPARγ-activating potential of the VLC fractions petroleum ether (PE), DCM-I and -II, and ethyl acetate extract (EtOAc) of the DCM extract from A. julibrissin no. 1a (a) and the DCM fractions D1-8 from Arisaema sp. no. 2a. (b). HEK293 cells, transiently transfected with expression plasmids for PPARα or PPARγ, luciferase reporter (tk-PPREx3-luc), and EGFP as internal control, were incubated for 18 h with 10 μg/mL of the indicated fraction, solvent vehicle (0.1% DMSO, negative control), and 50 nM GW7647 (PPARα agonist) or 5 μM troglitazone (PPARγ agonist) as positive control. Luciferase activity and fluorescence intensity were measured. Results are presented as mean ± SD (n = 4). Significantly different from the negative control (ANOVA), *P < 0.05; **P < 0.01.
Figure 3.
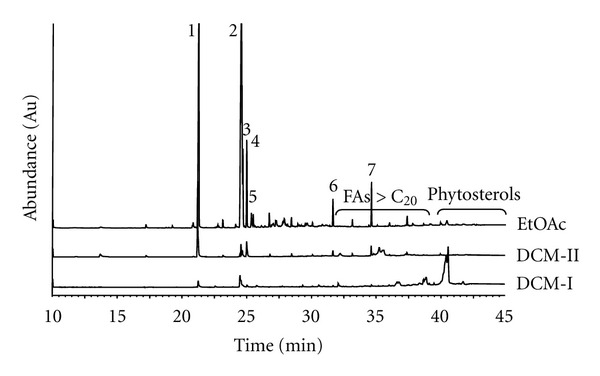
Comparative analysis by GC-MS of the DCM-I and -II and EtOAc VLC fractions (1 μL of 4 mg/mL in DCM) from the mother DCM extract of CHMs nos. 1a of A. julibrissin. FAMEs formed from the present FAs after derivatisation by TMSH were detected by GC-MS on a Zebron ZB-5 column (60 m × 0.25 mm, 0.25 μm) (Phenomenex) with a temperature gradient of 120°C-320°C in 40 min and 5 min hold; carrier gas: helium; flow rate: 1.7 mL/min. Ion source: EI 70 eV, 250°C. The following FAs were identified by GC-MS with a relative content (%) in each enriched extract given in the order DCMI; DCMII and EtOAc: (1) palmitic acid (2.9; 37; 23%), (2) linoleic acid (4.5; 1.0; 47%), (3) 9-octadecenoic acid (3.3; 5.1; 4.4%), (4) oleic acid (—; 3.8; 5.8%), (5) octadecanoic acid (3.3; 9.4; 2.2%), (6) docosanoic acid (—; 4.5; 1.7%), and (7) tetracosanoic acid (—; 2.3; 3.0%).
Figure 4.
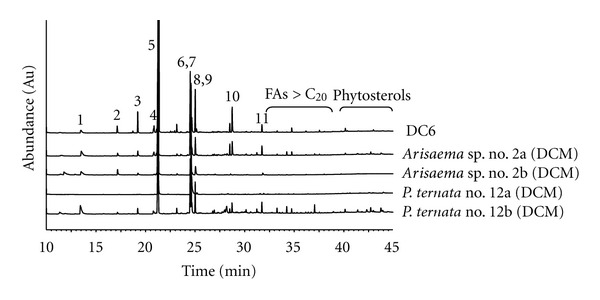
Comparative analysis by GC-MS of fraction D6 from the DCM extract of the CHM no. 2a from Arisaema sp. with DCM extracts from CHMs no. 2a and b of Arisaema sp. and no. 12a and b from P. ternata (1 μL of 4 mg/mL in DCM). FAMEs formed from the present FAs after derivatisation by TMSH were detected by GC-MS as in Figure 3. The following FAs were identified by GC-MS with a relative content (%) in each enriched extract given in the order of D6, CHM no. 2a, 2b, 12a and 12b: (1) nonanedioic acid (1.1; 4.1; 3.4;—; 3.8%), (2) tetradecanoic acid (1.0; 2.1; 1.0; —; 0.4%), (3) pentadecanoic acid (2.4; 1.5; 0.8;—; 0.9), (4) 9-hexadecenoic acid (1.5; 1.7; 0.4;—; 0.1%), (5) palmitic acid (61; 39; 74; 30; 37%) (6) linoleic acid (6.5; 8.9; 0.5; 36; 6.6%), (7) 9-octadecenoic acid (4.2; 8.2; 4.6; 17; 9.2%), (8) oleic acid (2.9; 1.6; 1.0;—; 2.6%), (9) octadecanoic acid (8.5; 4.5; 3.4; 2.6; 4.4%), (10) 13-phenyltridecanoic acid (3.4; 3.9; 0.5; 0.1; 2.6%), and (11) docosanoic acid (0.2; 0.9; 0.7;—; 0.3).
It became further evident that the content of the FAs present in “D6” was also high in the DCM extracts from CHMs from Arisaema sp. and P. ternata (Figure 4), especially of palmitic acid (30%–74%). The CHMs of T. terrestris and C. monnieri were previously described to contain FAs including palmitic acid, stearic acid, oleic acid, and linoleic acid [19, 32]. Besides common FAs, trihydroxy- and epoxy-hydroxy FAMEs and α-spinasterol-3-O-β-D-glucopyranoside were isolated from the MeOH extract of A. julibrissin no. 1a. Although mono- and dihydroxylated FAs are natural PPARγ agonists [40], trihydroxy- and epoxy-hydroxy FAMEs as well as their FA derivatives, identified by GC-MS [21], lacked PPARα and PPARγ activity in vitro in this study. Interestingly, chronic exposure to high levels of palmitic acid and stearic acid, identified as one of the main active principles in this study, are associated with lipotoxicity or insulin resistance, the main causative factor of type 2 diabetes and the metabolic syndrome [33, 39]. However, unsaturated free FAs, such as linoleic acid and oleic acid, do not promote insulin resistance [41]. In contrast, even a protective effect of oleic acid against palmitate-induced insulin resistance in L6 rat myotubes has been demonstrated [42]. Moreover, FAs have recently been identified as rather potent inhibitors of PTP1B, the main negative regulator of insulin and leptin signaling [43]. Thus, the impact of FAs on health and disease seems to highly depend on duration of exposure, concentration of the FA, or the degree of saturation.
4. Conclusion
In this study, extracts from CHMs were tested for a putative PPARα- or PPARγ-activating and NF-κB-inhibiting effect. Out of the fourteen plant species of which extracts were tested, 43% exhibited NF-κB inhibitory and 50% PPAR activating effects. The apolar PPAR active extracts and enriched fractions from flos and cortex of A. julibrissin and rhizomes of Arisaema sp. and P. ternata mainly contained fatty acids as PPAR-agonists, including palmitic acid, linoleic acid, oleic acid, and stearic acid. The outcome of this study contributes to the molecular understanding and explanation of some effects elicited by extracts of these three traditional Chinese herbs by revealing PPAR activation due to the present fatty acids.
Supplementary Material
Workflow of bioassayguided fractionation and isolation.
Acknowledgments
The authors thank Professor Ronald M. Evans for providing the PPRE luciferase reporter construct. They gratefully acknowledge the expert technical assistance of E. Geiger and J. Benedics. They thank for funding from the Sino-Austria Project, supported by the Austrian Federal Ministry of Science and Research and Federal Ministry of Health, Women and Youth. This project was also supported in part by the Austrian Science Fund [NFN S10704-B037].
Abbreviations
- ASE:
Accelerated solvent extractor
- BSTFA-TMCS:
N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) and trimethylchlorosilane
- CC:
Column chromatography
- CHM:
Chinese herbal medicine
- DCM:
Dichloromethane
- DMEM:
Dulbecco's modified Eagle's medium
- DMSO:
Dimethylsulfoxide
- EGFP:
Enhanced green fluorescent protein
- EI MS:
Electron ionization mass spectrometry
- EtOAc:
Ethylacetate
- FA:
Fatty acid
- FAME:
Fatty acid methyl ester
- FBS:
Fetal bovine serum
- GC-MS:
Gas chromatography mass spectrometry
- LPS:
Lipopolysaccharide
- MeOH:
Methanol
- NF-κB:
Nuclear factor κB
- NMR:
Nuclear magnetic spin resonance
- PE:
Petroleum ether
- PPAR:
Peroxisome proliferator-activated receptor
- TNF:
Tumor necrosis factor
- TMSH:
Trimethylsulfoniumhydroxide
- VLC:
Vacuum liquid chromatography.
References
- 1.Zhao Z, Liang Z, Chan K, et al. A unique issue in the standardization of Chinese materia medica: Processing. Planta Medica. 2010;76(17):1975–1986. doi: 10.1055/s-0030-1250522. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich M, Gibbons S. Ethnopharmacology in drug discovery: an analysis of its role and potential contribution. Journal of Pharmacy and Pharmacology. 2001;53(4):425–432. doi: 10.1211/0022357011775712. [DOI] [PubMed] [Google Scholar]
- 3.Cheung F. Made in China. Nature. 2011;480:S82–S83. doi: 10.1038/480S82a. [DOI] [PubMed] [Google Scholar]
- 4.Kinghorn AD, Pan L, Fletcher JN, Chai H. The relevance of higher plants in lead compound discovery programs. Journal of Natural Products. 2011;74(6):1539–1555. doi: 10.1021/np200391c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.communication Personal. CHMs in this study were selected in cooperation with Zhu Ming of the Medical Institute, Beijing University of Chinese Medicine & Pharmacology. Chaoyang District, Bei San Huan Dong Lu 11, Beijing 100029, China.
- 6.Singhuber J, Baburin I, Kahlig H, Urban E, Kopp B, Hering S. GABA(A) receptor modulators from Chinese herbal medicines traditionally applied against insomnia and anxiety. Phytomedicine. 2012;19(3-4):334–340. doi: 10.1016/j.phymed.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S, Hayden MS. New regulators of NF-κB in inflammation. Nature Reviews Immunology. 2008;8(11):837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 8.Bierhaus A, Schiekofer S, Schwaninger M, et al. Diabetes-associated sustained activation of the transcription factor nuclear factor-κB. Diabetes. 2001;50(12):2792–2808. doi: 10.2337/diabetes.50.12.2792. [DOI] [PubMed] [Google Scholar]
- 9.Salminen A, Lehtonen M, Suuronen T, Kaarniranta K, Huuskonen J. Terpenoids: Natural inhibitors of NF-κB signaling with anti-inflammatory and anticancer potential. Cellular and Molecular Life Sciences. 2008;65(19):2979–2999. doi: 10.1007/s00018-008-8103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glass CK. Going nuclear in metabolic and cardiovascular disease. Journal of Clinical Investigation. 2006;116(3):556–560. doi: 10.1172/JCI27913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chawta A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the x-files. Science. 2001;294(5548):1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- 12.Kershaw EE, Flier JS. Adipose tissue as an endocrine organ. Journal of Clinical Endocrinology and Metabolism. 2004;89(6):2548–2556. doi: 10.1210/jc.2004-0395. [DOI] [PubMed] [Google Scholar]
- 13.Delerive P, De Bosscher K, Besnard S, et al. Peroxisome proliferator-activated receptor α negatively regulates the vascular inflammatory gene response by negative cross-talk with transcription factors NF-κB and AP-1. Journal of Biological Chemistry. 1999;274(45):32048–32054. doi: 10.1074/jbc.274.45.32048. [DOI] [PubMed] [Google Scholar]
- 14.Daynes RA, Jones DC. Emerging roles of PPARs in inflammation and immunity. Nature Reviews Immunology. 2002;2(10):748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- 15.Shearer BG, Billin AN. The next generation of PPAR drugs: do we have the tools to find them? Biochimica et Biophysica Acta. 2007;1771(8):1082–1093. doi: 10.1016/j.bbalip.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Huang THW, Kota BP, Razmovski V, Roufogalis BD. Herbal or natural medicines as modulators of peroxisome proliferator-activated receptors and related nuclear receptors for therapy of metabolic syndrome. Basic and Clinical Pharmacology and Toxicology. 2005;96(1):3–14. doi: 10.1111/j.1742-7843.2005.pto960102.x. [DOI] [PubMed] [Google Scholar]
- 17.Berger JP, Petro AE, Macnaul KL, et al. Distinct properties and advantages of a novel peroxisome proliferator-activated protein γ selective modulator. Molecular Endocrinology. 2003;17(4):662–676. doi: 10.1210/me.2002-0217. [DOI] [PubMed] [Google Scholar]
- 18.Cavender MA, Lincoff AM. Therapeutic potential of aleglitazar, a new dual PPAR-αγ agonist: implications for cardiovascular disease in patients with diabetes mellitus. American Journal of Cardiovascular Drugs. 2010;10(4):209–216. doi: 10.2165/11539500-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 19.Bensky D, Clavey S, Stöger E, Gamble A. Chinese Herbal Medicine Materia Medica. Seattle, Wash, USA: Eastland Press; 2004. [Google Scholar]
- 20.Kuang L, Zhang K. Pharmacopoeia of the People's Republic of China. Beijing, China: People's Medical Publishing House; 2010. [Google Scholar]
- 21.Hamberg M. Regio- and stereochemical analysis of trihydroxyoctadecenoic acids derived from linoleic acid 9- and 13-hydroperoxides. Lipids. 1991;26(6):407–415. doi: 10.1007/BF02536065. [DOI] [PubMed] [Google Scholar]
- 22.Peng C, Bodenhausen G, Qiu S, et al. Computer-assisted structure elucidation: application of CISOC-SES to the resonance assignment and structure generation of betulinic acid. Magnetic Resonance in Chemistry. 1998;36(4):267–278. [Google Scholar]
- 23.Sholichin M, Yamasaki K, Kasaj R, Tanaka O. 13C Nuclear magnetic resonance of lupane-type triterpenes, lupeol, betulin and betulinic acid. Chemical and Pharmaceutical Bulletin. 1980;28:1006–1008. [Google Scholar]
- 24.Ishi H, Nakamura M, Seo S, Tori K, Tozyo T, Yoshimura Y. Isolation, characterization and nuclear magnetic resonance spectra of new saponins from the roots of Bupleurum falcatum L. Chemical and Pharmaceutical Bulletin. 1980;28:2367–2383. [Google Scholar]
- 25.Tandon M, Shukla YN, Thakur RS. Steroid glycosides from Asparagus adscendens. Phytochemistry. 1990;29(9):2957–2959. [Google Scholar]
- 26.Fakhrudin N, Ladurner A, Atanasov AG, et al. Computer-aided discovery, validation, and mechanistic characterization of novel neolignan activators of peroxisome proliferator-activated receptor γ . Molecular Pharmacology. 2010;77(4):559–566. doi: 10.1124/mol.109.062141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baek MY, Cho JG, Lee DY, Ahn EM, Jeong TS, Baek NI. Isolation of triterpenoids from the stem bark of Albizia julibrissin and their inhibition activity on ACAT-1 and ACAT-2. Journal of Applied Biological Chemistry. 2010;53(3):310–315. [Google Scholar]
- 28.Rabi T, Shukla S, Gupta S. Betulinic acid suppresses constitutive and TNFα-induced NF-κB activation and induces apoptosis in human prostate carcinoma PC-3 cells. Molecular Carcinogenesis. 2008;47(12):964–973. doi: 10.1002/mc.20447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun F, Xie ML, Xue J, Wang HB. Osthol regulates hepatic PPARα-mediated lipogenic gene expression in alcoholic fatty liver murine. Phytomedicine. 2010;17(8-9):669–673. doi: 10.1016/j.phymed.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Xie ML, Xue J, Gu ZL. Osthole regulates enzyme protein expression of CYP7A1 and DGAT2 via activation of PPARalpha/gamma in fat milk-induced fatty liver rats. Journal of Asian natural products research. 2008;10(7-8):807–812. doi: 10.1080/10286020802102303. [DOI] [PubMed] [Google Scholar]
- 31.Liao PC, Chien SC, Ho CL, et al. Osthole regulates inflammatory mediator expression through modulating NF-KB, mitogen-activated protein kinases, protein kinase C, and reactive oxygen species. Journal of Agricultural and Food Chemistry. 2010;58(19):10445–10451. doi: 10.1021/jf102812t. [DOI] [PubMed] [Google Scholar]
- 32.Jiang E, Li H, Yang S. Effect of gross saponin Tribulus terrestris on NF-kappa B in PC12 cells injured by oxidative stress and its mechanisms. Jilin Daxue Xuebao, Yixueban. 2010;36:1035–1038. [Google Scholar]
- 33.Shi C, Qu W, Gao J. Effects of saponin from Tribulus terrestris on gene expression of ICAM-1, VCAM-1, PPARalpha and PPARgamma in artery vessels of atherosclerotic rats. Tianran Chanwu Yanjiu Yu Kaifa. 2009;36:761–765. [Google Scholar]
- 34.Kim YJ, Shin YO, Ha YW, Lee S, Oh JK, Kim YS. Anti-obesity effect of Pinellia ternata extract in Zucker rats. Biological and Pharmaceutical Bulletin. 2006;29(6):1278–1281. doi: 10.1248/bpb.29.1278. [DOI] [PubMed] [Google Scholar]
- 35.Yang DJ, Chang YY, Hsu CL, et al. Antiobesity and hypolipidemic effects of polyphenol-rich longan (dimocarpus longans lour.) flower water extract in hypercaloric-dietary rats. Journal of Agricultural and Food Chemistry. 2010;58(3):2020–2027. doi: 10.1021/jf903355q. [DOI] [PubMed] [Google Scholar]
- 36.Chao CY, Huang CJ. In vitro activation of peroxisome proliferator activated receptor α by some extracts from food materials. Yaowu Shipin Fenxi. 2008;16:62–69. [Google Scholar]
- 37.Meija J, Soukup VG. Phenyl-terminated fatty acids in seeds of various aroids. Phytochemistry. 2004;65(15):2229–2237. doi: 10.1016/j.phytochem.2004.06.033. [DOI] [PubMed] [Google Scholar]
- 38.Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(6):2160–2164. doi: 10.1073/pnas.90.6.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfrum C, Borrmann CM, Borchers T, Spener F. Fatty acids and hypolipidemic drugs regulate peroxisome proliferator-activated receptors alpha- and gamma-mediated gene expression via liver fatty acid binding protein: a signaling path to the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:2323–2328. doi: 10.1073/pnas.051619898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh T, Fairall L, Amin K, et al. Structural basis for the activation of PPARγ by oxidized fatty acids. Nature Structural and Molecular Biology. 2008;15(9):924–931. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirabara SM, Curi R, Maechler P. Saturated fatty acid-induced insulin resistance is associated with mitochondrial dysfunction in skeletal muscle cells. Journal of Cellular Physiology. 2010;222(1):187–194. doi: 10.1002/jcp.21936. [DOI] [PubMed] [Google Scholar]
- 42.Gao D, Griffiths HR, Bailey CJ. Oleate protects against palmitate-induced insulin resistance in L6 myotubes. British Journal of Nutrition. 2009;102(11):1557–1563. doi: 10.1017/S0007114509990948. [DOI] [PubMed] [Google Scholar]
- 43.Steinmann D, Baumgartner RR, Heiss EH, et al. Bioguided isolation of (9Z)-Octadec-9-enoic acid from Phellodendron amurense Rupr. and identification of fatty acids as PTP1B inhibitors. Planta Medica. 2012;78(3):219–224. doi: 10.1055/s-0031-1280377. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Workflow of bioassayguided fractionation and isolation.


