Abstract
The high-throughput structure determination pipelines developed by structural genomics programs offer a unique opportunity for data mining. One important question is how protein properties derived from a primary sequence correlate with the protein’s propensity to yield X-ray quality crystals (crystallizability) and 3D X-ray structures. A set of protein properties were computed for over 1,300 proteins that expressed well but were insoluble, and for ~720 unique proteins that resulted in X-ray structures. The correlation of the protein’s iso-electric point and grand average hydropathy (GRAVY) with crystallizability was analyzed for full length and domain constructs of protein targets. In a second step, several additional properties that can be calculated from the protein sequence were added and evaluated. Using statistical analyses we have identified a set of the attributes correlating with a protein’s propensity to crystallize and implemented a Support Vector Machine (SVM) classifier based on these. We have created applications to analyze and provide optimal boundary information for query sequences and to visualize the data. These tools are available via the web site http://bioinformatics.anl.gov/cgi-bin/tools/pdpredictor.
Keywords: Bioinformatics, Data mining, Protein crystallization, Crystallizability
Introduction
Protein X-ray crystallography requires high-quality single crystals for structure determination. It has been known for a long time that not all proteins can be crystallized, and therefore are not suitable for structure determination using X-ray crystallography. Identifying such proteins early in the structure determination process can save a substantial effort. Moreover, understanding protein propensities for crystallization can help to identify more suitable targets and design better constructs for structure determination.
To improve crystallizability of recalcitrant proteins a number of experimental approaches have been developed. These included limited [1, 2] or in situ proteolysis [3] to identify and cleave off poorly ordered regions, mutagenesis to reduce surface entropy [4], reductive methylation to promote crystallization [5], or the use of sequence orthologs [6] or truncations to select better protein or domain [7].
The structural genomics programs developed high throughput pipelines for structure determination from gene to 3-D structure and processed a large number of protein samples. Many selected proteins failed at various steps of the pipeline, and it was quickly realized that the attrition rate from a selected protein target to a successful PDB deposit is more than 90% (see the MCSG website for Protein Structure Initiative (PSI) statistics at http://www.mcsg.anl.gov). However, because these proteins were treated using standard protocols and selected proteins have low sequence similarities, the data can be used to extract protein properties affecting crystallizability.
Several approaches have been reported to predict the protein amenability to crystallization given an amino acid sequence [7–14]. The analyzed data sets range from a few hundred sequences to an entire protein set deposited in the Protein Data Bank (PDB). Prediction methods range from simple calculations based on iso-electric point (pI) and grand average hydropathy (GRAVY), or di- and tri-peptide profiles, to sophisticated methods where several predictions including secondary structure, disorder, and residue conservation are included. A comparison of the T. maritima proteome and crystallized proteins from this organism identified pI and GRAVY as one of the few key sequence-derived attributes affecting the successful outcome of crystallization effort [9]. Three clusters were observed on pI vs. GRAVY plots with differing proportions of crystallized proteins. A larger dataset was used to develop the OB-score including structures available in the PDB and UniRef50 database [12]. The SECRET sequence-based predictor of crystallizability [14] uses mono-, di-, and tri-peptide frequencies, and amino acid groups according to different hydrophobicity scales in a two-layer classifier, where the output of the first Support Vector Machine (SVM) classifier is used in a second step Naïve Bayes (NB) classifier. The predictor achieves a maximum of 65 and 69% accuracy for positive and negative classes, respectively. The CRYSTALP method, analyzing the same dataset as in the SECRET method, uses a reduced set of properties (46 vs. 103) with increased accuracy (77%). However, the CRYSTALP and SECRET methods have protein length limitations (46–200 amino acids). In the case of the Pxs predictor, the probability of crystallization was identified from biochemically well-behaved proteins from which crystal structures were obtained using properties such as the fraction of disordered residues, the mean side-chain entropy of predicted exposed residues, the fraction of predicted buried glycines and phenylalanine frequency [15]. Even more sophistication is provided by XtalPred [7], where predicted secondary structure, disordered regions, residue conservations are also considered. Precalculated values are also available on the XtalPred website for completed microbial genomes providing a very important resource for structural genomics pipelines.
Because the experimental setting may also affect the analysis we decided to use a smaller but self-consistent set of sequences from the Midwest Center for Structural Genomics (MCSG) protein target list. All selected proteins were processed using this same set of experimental steps and conditions.
We applied statistical and data mining techniques to extract the most important attributes influencing protein propensity to crystallize. We then used the generated metrics for selecting targets more suitable for crystallization and for selection of better full lengths protein targets and for the design of domain constructs.
Materials and methods
The selection of the data set
The protein sequence data set used in this study comprised of 1,346 proteins that expressed in E. coli but were not soluble (MCSG-INSOLUBLE) and 723 proteins that passed through all steps of the pipeline and resulted in PDB deposits (MCSG-PDB) selected from the MCSG database. All proteins were expressed using N-terminal His6-tag containing vectors (pMCSG7) and were processed according to the MCSG standard operating procedures (SOPs). The MCSG Laboratory Information Management System (LIMS) database was used to select insoluble targets and those deposited into the PDB. In order to reduce sequence redundancy, the sequences were clustered using cd-hit [16] and the 30% identity class was used in this study. The targets were obtained from 130 species according to a selection process described earlier [17]. Briefly, proteins that belong to a given family are identified from the MCSG reagent genomes using the HMMER package [18–22]. The boundaries of the sub-sequence defined by the Hidden Markov Model (HMM) are extended until disordered or transmembrane regions are encountered using DISOPRED and TMHMM for prediction, respectively [23, 24]. In addition, targets are further filtered using BLAST for the identification of PDB-similarity [25].
Calculations
The OB-score was calculated by the software provided by the authors [12]. The program utilizes a Z-score matrix derived from the clustering of pI and GRAVY values calculated for a reduced set of PDB entries and UNIPROT sequences [12]. We have previously developed methods for the calculation of pI with fine granularity (pKa variants) and in good agreement with the predicted values observed in two-dimensional gel electrophoresis-based proteomics [26]. In this study we used our software (http://gelbank.anl.gov/cgi-bin/proteomes/titration_seq.pl) for the calculation pI and GRAVY and have constructed a Z-score matrix derived from the comparison of the MCSG-INSOLUBLE and MCSG-PDB datasets (see above).
Additional amino acid attributes were selected for data mining purposes from the AAindex database [27]. The nearly 500 amino acid attributes from the AAindex were normalized and clustered in order to reduce them to a set of 30 attribute classes using a Perl implementation of the Kohonen Maps (AI::NeuralNet::SOM). Kohonen maps (Self-organizing maps) can be used to produce two-dimensional representation of the input space of the training samples while preserving their topological properties [28]. Two representatives of each class were used to calculate sequence properties: (1) the amino acid composition was used to derive an average value, and (2) a 7 amino acid-long sliding window was used to define an overall minimum and maximum for a given target. In addition, amino acid and di-peptide frequencies were calculated for every target. Overall more than 400 attributes were calculated for every sequence in the MCSG-INSOLUBLE and MCSG-PDB sets. A subset of these attributes was selected using Student’s t-test. This reduced data set was used in data mining employing a Support Vector Machine (SVM) approach with a Gaussian kernel (the tolerance was kept at 0.001, the complexity factor was 2.25 ± 0.12, and active learning was enabled in the model generation).
Web applications
Web applications were built for a number of steps: 2-dimensional (2D) binning of matrices, display of 2D matrices, and Z-score matrix generation using an unbalanced set of inputs with Monte-Carlo sampling (http://bioinformatics.anl.gov/cgi-bin/tools/pdpredictor; supplementary information). The web applications are hosted on a Windows 2003 server running IIS6.0. The calculations were performed on a local Linux cluster. The Perl GD library was used for the visualization in the web applications. The Oracle Data Mining package (10gR2) was used for data mining.
Results
The construction of the data sets
Currently, the MCSG (and other Protein Structure Initiative centers) protein network targets are assigned based on the protein sequence analysis using Hidden Markov Models (HMMs). Each center selects several orthologs using the given HMM profile from a set of reagent genomes available for cloning. For each protein target several constructs are put into the pipeline: the full length protein sequence (if no trans-membrane domain or signal peptides are detected), and extended versions of the domain determined by the HMM based on trans-membrane, low complexity, and disordered region predictions [17]. In addition, prior to cloning targets are screened for PDB similarity and only those with less than 30% sequence identity are processed. The targets are further filtered by percent methionine content and molecular weight, with minimum values of 0.7% and 10 kDa, respectively. Most of the targets selected for this study were cloned into the pMCSG7 vector, adding a N-terminal His6-tag which enables high throughput affinity purification [29]. The targets are screened for expression and solubility in small scale and only soluble proteins are purified in large scale and set up for crystallization. The MCSG Laboratory Information Management System (LIMS) was used to select two datasets: (a) proteins that were insoluble in the small-scale screening process and (b) those deposited into the PDB from the MCSG pipeline. In the MCSG pipeline, orthologous targets are used, generating sequence classes with high similarity. Therefore both the insoluble and PDB entries were clustered at 30% using cd-hit in order to reduce redundancy, resulting in the final data sets used in this study, MCSG-INSOLUBLE (1,346 members) and MCSG-PDB (723 members) (Figure S1). The clustering retained more than 85% of all MCSG deposits, but only ~50% of the insoluble targets due to the orthologous target processing approach. The molecular weight of all targets ranges from 5 to 200 kDa with an average MW of 26.5 kDa for MCSG-INSOLUBLE and 23.3 kDa for MCSG-PDB, with a significant difference between the two classes (P < 0.001; Figure S2).
The clones derived from nearly 130 species with almost complete overlap between MCSG-INSOLUBLE and MCSG-PDB datasets. Grouping all targets by species shows a wide range of variations for soluble clones amenable for large-scale purification. When the number of clones was normalized for those species with at least 50 constructs tested, on average Xylella fastidiosa Temecula1, Listeria innocua, and ATCC 19718 yielded the least, while Staphylococcus aureus subsp. aureus N315, Aquifex aeolicus, and Bacillus subtilis subsp. subtilis str. 168 yielded the most soluble clones that were further processed in the MCSG pipeline. Consequently, constructs from L. innocua, B. henselae str. Houston-1, and X. fastidiosa Temecula1 yielded the least MCSG structures, while S. aureus subsp. aureus N315, S. pneumoniae TIGR4, B. subtilis subsp. subtilis str. 168, and A. aeolicus were the most productive species in this analysis (Figure S3). For the majority of the species there is a positive trend between the percent soluble clones and the percent PDB entries. Poor correlation was found for the following species: Streptococcus pneumoniae TIGR4, Saccharomyces cerevisiae, Nitrosomonas europaea ATCC 19718, Bacillus subtilis subsp. subtilis str. 168, Chlamydomonas reinhardtii, and Caenorhabditis elegans.
The construction of an initial Z-score matrix was based on pI and GRAVY index and its use in target selection and subregion design
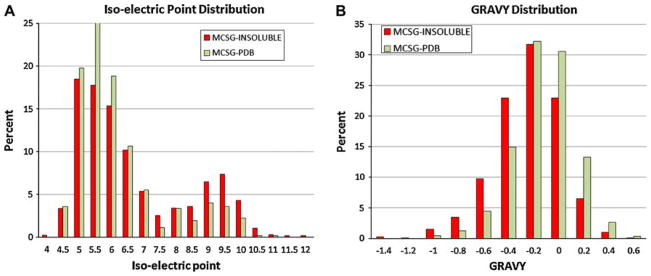
We explored the applicability of OB-score for the analysis of protein sequences using the MCSG-INSOLUBLE and MCSG-PDB data sets. We calculated the pI, GRAVY index, and OB-score using the OB_score program [12]. Similarly to previously reported values, the MCSG pipeline had better success with more acidic and slightly hydrophobic targets (Fig. 1). The OB-score distribution was similar to that reported earlier [12], although we observed less discrimination when applied to the MCSG targets. Since the OB-score was constructed from a large dataset, we wanted to see if the MCSG pipeline data could be used to generate an MCSG pipeline-specific Z-score matrix.
Figure 1.
Iso-electric point and GRAVY distribution of MCSG targets. The pI (a) and hydrophobicity (b) of MCSG-INSOLUBLE (dark) and MCSG-PDB (light) targets were calculated and binned according to the OB-score matrix bin values [12]
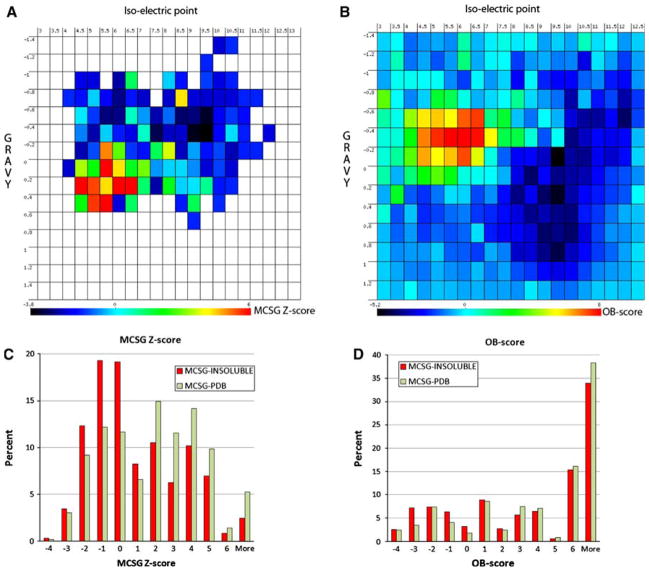
We used reported amino acid attributes for the calculation of GRAVY, while slightly different pKa values for the calculations of pI. Two-dimensional gel electrophoresis-based proteomics can resolve proteins that differ slightly in pI [30]. This method allows developing approaches more accurately predicting “true” pI [26]. We recalculated the pI and GRAVY using this new approach (see Methods section). While the pI values correlated well for the majority of the protein sequences, they differed around neutral pI values (Figure S4). The sizes of the MCSG-INSOLUBLE and MCSG-PDB data sets are different (~1,346 vs. ~723). We have performed a repeated random sub-sampling validation using a 60/40 split of the non-redundant data. Briefly, the 30% non-redundant data-set was split randomly into two sets at a 60 to 40 ratio. The 60% set was used to build a Z-score matrix, while the 40% set was used for validation. The MCSG-INSOLUBLE dataset is larger and was used to generate average and standard error information for each pI/GRAVY bin via a Monte-Carlo sampling (1,000 rounds). The MCSG-PDB data set was used to compare with the above matrix in order to construct an MCSG-specific Z-score matrix (Fig. 2). We used the same binning as reported for the OB-score matrix. When compared, the MCSG Z-score matrix is more compact, with data points occupying less than ¼ of the matrix (Fig. 2a), while every bin in the Z-score matrix of the OB-score program has a value due to the fact that much larger datasets (UniRef50 and PDB) were used in their calculation (Fig. 2b). The missing values in the MCSG Z-score matrix can be approximated by 0.
Figure 2.
Comparison of the Z-score matrix derived from MCSG targets and the OB-score matrix. A web application was built for binning two-dimensional data (a) and for the display of two-dimensional matrices (b). The OB-score distribution is shown for pI 3–13 and GRAVY −1.4 to 1.4 as reported earlier by Overton and Barton [12]. The selected insoluble (dark) and ‘In PDB’ targets (light) were binned according to the MCSG Z-score (c) and the OB-score (d)
We calculated the OB-score and the MCSG Z-score for the 40% set was used for validation. The MCSG Z-score matrix generation and its evaluation were repeated 5 times. We compared the Z-scores calculated by the OB_score program and by our method (Figure S5). The calculated Z-scores were binned (Fig. 2c, d) and the area under the receiver operating characteristic (ROC) curve was calculated (AUC-ROC; Table 1). The OB-score method returned a marginal difference from a random guessing (~52%), while the MCSG Z-score showed a modest performance (~61%). Given the simple calculation of the MCSG Z-score it allows the scanning of tens of thousands of sequences within a short time on a single processor.
Table 1.
The AUC-ROC of the OB-score and MCSG Z-score predictions
| Method | AUC-ROC (%) | SD (%) |
|---|---|---|
| OB-score | 51.80 | 1.20 |
| MCSG Z-score | 60.80 | 1.00 |
A repeated random sub-sampling validation was performed using a 60/40 split of the non-redundant data (30% cutoff). The 60% set was used to generate a Z-score matrix and the 40% set was used to calculate the OB-score and MCSG Z-scores. The AUC-ROC was calculated for both Z-score methods by varying the decision threshold
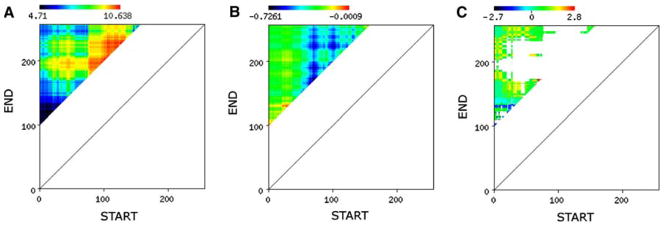
We also explored how Z-score calculations could aid in designs of target truncations. There are several cases when none of the orthologs of a given target selected for the MCSG pipeline are soluble requiring the design of subregions of the protein targets. The main question is how to select or design the “protein” or “domain” to obtain high quality structure? Given an average target length of 230 amino acids there are nearly 8,500 possible truncations of a sequence with a minimum of 100 amino acid length. The calculation of pI, GRAVY, and Z-score for all possible subregions of at least 100 amino acids in length of an example protein sequence is shown in Fig. 3. Subregions of a protein target can vary wildly in terms of pI and GRAVY values. The MCSG Z-score not only reveals areas with tendencies for crystallizability and for insolubilty, albeit at low accuracies, but also areas corresponding to constructs with pI/GRAVY combinations that have never been processed in the MCSG pipeline. This is a distinct feature of the MCSG Z-score.
Figure 3.
Subregion design. A web application was built for calculating the pI, GRAVY, and the corresponding MCSG derived Z-score for all possible subregions of an input sequence. The resulting matrix is displayed for pI (a), GRAVY (b) and the MCSG Z-score (c) using a predefined color scale
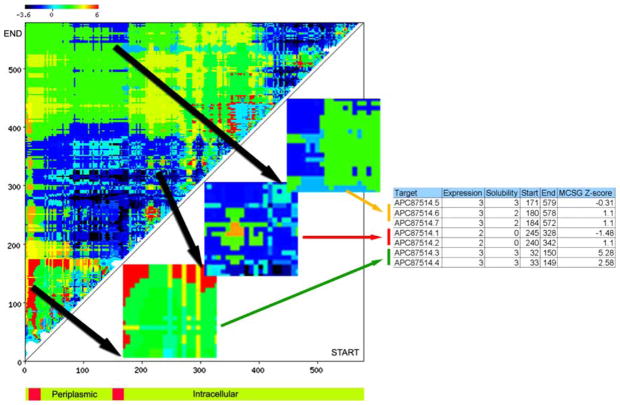
The example in Fig. 3 shows a variation of pI between 4.7 and 10.6, and a smaller variation in GRAVY between −0.7 and 0. The MCSG Z-score reveals a large “unknown” space in terms of the pI-GRAVY combination (not observed thus far at the MCSG). Based on this data a design process could be guided by the MCSG Z-score matrix. In the case of one of the B. subtilis targets in the MCSG pipeline, the two-component sensor histidine kinase (APC87514; gi|221310851), the full-length protein was not selected, but several subregions were designed and tested in the pipeline (Fig. 4). This protein has two predicted transmembrane α-helices with a short periplasmic region and a long cytoplasmic tail as predicted by Phobius [31]. The cytoplasmic tail contains a his-tidine kinase A dimerization/phosphoacceptor domain (pfam00512; residues 348–400) and a histidine kinase-like ATPase domain (pfam02518; 468–571). We have designed two sets of constructs in the predicted periplasmic region (around residues 33–150), three longer constructs (around residues 171–579) and two shorter constructs (around residues 240–342) in the predicted cytoplasmic tail. All designs expressed well and while the periplasmic constructs were soluble, both cytoplasmic constructs were insoluble. We have obtained a structure for one of the periplasmic constructs (APC87514.3, PDB:3CWF), revealing a novel PAS-domain (PhoR) [32]. The soluble proteins for the longer cytoplasmic tail designs are still in crystallization trials.
Figure 4.
Constructs of a two-component sensor histidine kinase. Several constructs of a two-component sensor histidine kinase from Bacillus subtilis subsp. subtilis str. 168 (gi|221310851) were designed and tested in the pipeline. The expression and solubility data of 3 selected subregions are shown. The bottom schematics depicts the topology of the protein as predicted by Phobius [31]
It is important to note that the MCSG data set does not contain targets with predicted transmembrane regions, while earlier studies contrasted PDB entries with full pro-teomes with an average of 30% membrane proteins (for example the T. maritima or the UniRef50 sequences; [9, 12]). The MCSG Z-score (or simply the binned pI-GRAVY distribution) can distinguish targets that have not been successfully crystallized in the MCSG pipeline before, but is insensitive to disordered regions or can even incorrectly identify them as favorable regions (Fig. 4 matrix at the N-terminal region).
Enhancing Z-score matrix using an extended set of protein sequence properties
The approach described above uses only two descriptors, the pI and GRAVY, of a protein sequence to develop a Z-score for predicting crystallizability of a given sequence or an array of subregions of a given sequence. We have tested how other sequence-derived properties affect the success in obtaining crystal structure using the MCSG pipeline. We used the AAindex database of biochemical and physicochemical properties of amino acids [33] as a starting point. The nearly 500 available attributes were first normalized to a 0–1 scale. Since the 500 attributes are related, they can be clustered based on similarities. Here we reduced the 500 attributes to a set of an arbitrarily chosen 30 attribute classes using Kohonen Maps and two representatives of each class were selected in our analysis (Figure S6). We have used the same data set as described for the construction of the MCSG Z-score matrix for this analysis (MCSG-INSOLUBLE and MCSG-PDB). For each sequence the amino acid composition was calculated and overall protein property was derived using the given amino acid attribute. In addition to calculating an average value for each attribute as derived from the amino acid composition, we have calculated local minimum and maximum values using a 7 amino acid size sliding-window for every sequence. Furthermore, we also calculated the molecular weight, pI, and amino acid distribution (mono- and dipeptide frequencies). The GRAVY was included as one of the attributes in the 30 property classes. The di-peptide frequencies alone generate 400 values for a given sequence in addition to nearly 200 attributes derived from the 60 amino acid properties—two from each class—with minimum, average, and maximum calculations.
The calculated attributes were analyzed using Student’s t-test and bin values were calculated for each attribute guided by standard deviations (σ) within the data sets (using 2σ in order to map values reasonably well around the means). For every attribute, 11 bins were created and the MCSG-INSOLUBLE and MCSG-PDB data sets were further analyzed using a Monte Carlo sampling in order to equalize the two data sets. The MCSG-INSOLUBLE and MCSG-PDB data sets were sampled separately 1,000 times with a sample size of 500. The results were then used to define attribute correlation (or anti-correlation) with the structure determination success rate using overall discriminatory power. The Student’s t-test values were in good agreement with the attribute importance derived from the above method. The top 20 attributes that correlate crystallizability are listed in Table 2. Most of the attributes originate from the AAindex1 database and are related to protein structure information or the presence or absence of transmembrane helices. Some represent average properties for a protein sequence, such as average volume of buried residue [34] or averaged turn propensities in a transmem-brane helix [35], while some describe local minimum or maximum values, such as the turn propensity scale for transmembrane helices [36] or normalized frequency of alpha-helix in all-alpha class [37]. Interestingly, while the pI is a distinguishing factor between the MCSG-INSOLUBLE and MCSG-PDB data sets, the GRAVY of a protein sequence was trailing behind the top attributes listed in Table 2. Some amino acid frequencies also correlate well with crystallizabillity (C, E, H, M, N, S, and Y frequencies) (Table 3).
Table 2.
Attributes identified for data mining
| Attribute | Description | Reference |
|---|---|---|
| CHOC750101FULL | Average volume of buried residue | [34] |
| CHOP780215FULL | Frequency of the 4th residue in turn | [42] |
| MONM990101MAX | Turn propensity scale for transmembrane helices | [36] |
| MONM990201FULL | Averaged turn propensities in a transmembrane helix | [35] |
| MUNV940105MAX | Free energy in beta-strand region | [43] |
| PALJ810107MIN | Normalized frequency of alpha-helix in all-alpha class | [37] |
| PONP800101MIN | Surrounding hydrophobicity in folded form | [37, 46] |
| QIAN880116MAX | Weights for beta-sheet at the window position of −4 | [44] |
| QIAN880138MIN | Weights for coil at the window position of 5 | [44] |
| RACS820101FULL | Average relative fractional occurrence in A0(i) | [47] |
| RICJ880104FULL | Relative preference value at N1 | [45] |
| TANS770104MAX | Normalized frequency of chain reversal R | [45, 48] |
| pI | Isoelectric point | Methods |
| C,E,H,M,N,S,Y | Amino acid frequencies |
The AAindex1 database was used to calculate minimum, maximum, and average values for the MCSG-INSOLUBLE and MCSG-PDB data sets. The attributes were ranked using a Student’s t-test. The top amino acid attributes are shown. In addition the iso-electric point and certain amino acid frequencies showed discriminatory power. The attributes listed here were used in data mining
Table 3.
Accuracy of the SVM prediction
| Target value | Total actuals | Correctly predicted (%) | Cost |
|---|---|---|---|
| Insoluble | 549 | 56.50 | 239 |
| In PDB | 290 | 74.80 | 73 |
The attributes identified in Table 2 were used to calculate properties for the MCSG-INSOLUBLE and the MCSG-PDB data sets. The generated matrix was used in a clustering using a SVM approach splitting the entries 60–40%. The 60% portion was used for training while the 40% portion for evaluating the accuracy of the prediction. The SVM model building and validation were repeated five times. The insoluble proteins were predicted at 56% while the PDB deposits were predicted at 75% accuracy
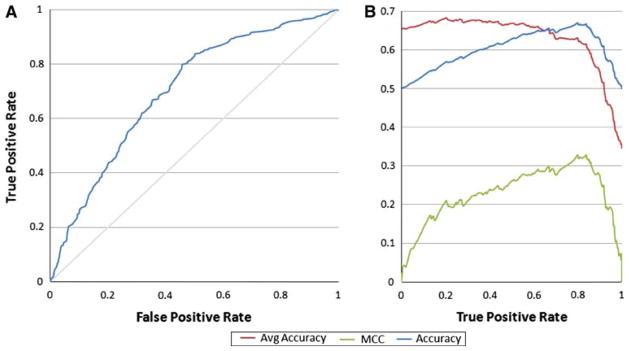
In the next step we used these attributes in a Support Vector Machine (SVM) classifier [38] using Oracle’s implementation. The sample set was divided up and 60% was used for training, while the rest was used for testing purposes and the classification was performed using a Gaussian kernel function. This method predicted the positive class (PDB deposits) at 74.8% accuracy and the negative class (insoluble proteins) at 56.47% accuracy. The area under the receiver operating characteristic (ROC) curve (AUC-ROC) was 68.3 ± 1% (random prediction would provide 50%, while perfect prediction would result in 100% AUC-ROC). In order to measure the quality of the binary classification we also calculated the Matthews Correlation Coefficient (MCC, [39]) as follows:
The MCC was maximal at the average accuracy reported (around 0.3). The MCC describes the correlation between the observed and predicted binary classifications (1 is a perfect prediction while 0 is a random prediction (Fig. 5).
Figure 5.
The Support Vector Machine approach. The amino acid attributes selected above were used to calculate protein sequence properties for a balanced set of insoluble targets and those targets deposited into the PDB. A repeated random sub-sampling validation was performed using a 60/40 split of the non-redundant data. The SVM was trained with 60% of the dataset and the remainder was used for testing. The true positive and false positive rate is shown in (a) with a 68% AUC-ROC. The MCC was calculated and displayed along the accuracy and average accuracy in relation to the true positive rate (b)
The use of SVM model for generating target truncations is feasible but computationally expensive. Nevertheless, two-dimensional matrix for a protein sequence can be created similarly to the MCSG Z-score method as presented above (not shown).
Discussion and conclusion
The PSI structural genomics pipelines not only generate a large number of novel structures but also provide experimental data (both successes and failures) that can be utilized for data mining activities. The prediction methods are very important in guiding target selection and prioritization in order to lower the attrition rates in the SG pipelines. There are several methods available to estimate a given target’s crystallizability. Previous studies identified the importance of pI, GRAVY index, and several other attributes, such as amino acid composition, predicted secondary structure, disordered regions, and residue conservations [7, 12, 14, 15, 40, 41] that correlate with success in crystallization.
Here we describe a method that is based on a single center’s experimental information. While most predictors derive crystallization propensity purely from the sequence information, we believe that the Standard Operating Procedures (SOPs) used in every step of the pipeline also affects crystallizability, ranging from expression vectors, cell growth and induction protocols, purification protocols to the application of crystallization screens. The SG large centers have applied the SOPs in the crystallization of thousands of proteins and provide valuable metadata for every target at every step of the pipeline. First we explored the applicability of a Z-score matrix constructed similarly to that of the OB-score using the MCSG metadata. We found that the OB-scores did not perform well and the MCSG Z-score matrix showed a modest performance (~61% AUC-ROC), but the sparse Z-score matrix can be exploited in the design of truncations or for the identification of targets that are drastically different than that part of the more than 30,000 targets processed in the pipeline. We have sought to identify additional amino acid attributes from the AAindex database that could be used for data analyses. The relevant attributes were mostly related to amino acid properties derived from previous structural biology efforts. Using a SVM method including these additional attributes we predicted the positive class, proteins that result in the crystal structures deposited into the PDB, at 75%, while the negative class, proteins that fail to express in soluble form, at 56% accuracy. The AUC-ROC curve was 68% providing substantial improvement over random selection. This method is now being used in designing domain constructs at MCSG.
It is important to note that all predictors use slightly different data sets for comparison: on one end full proteomes, while on the other end soluble proteins are contrasted with PDB deposits. In this study we compared targets that were selected by our bioinformatics pipeline, eliminating a number of factors others studied and found relevant, such as highly hydrophobic and disordered regions. The contrasted data sets were set up as insoluble versus X-ray quality crystal producing target pools in hope of identifying these cases for the incoming targets. The MCSG Z-score can be useful for quick identification of amenable truncation designs, while the more computationally intensive SVM approach for the design of truncations of targets with high importance.
We have developed several web applications for generating and displaying two-dimensional matrices, and we have enabled a publicly available web application for the prediction of the MCSG Z-score for a list of input sequences. These tools are accessible via the MCSG web site (http://bioinformatics.anl.gov/cgi-bin/tools/pdpredictor). Currently we are investigating the incorporation and the effect of attributes used for the XtalPred server [7].
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health grant GM074942 and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Abbreviations
- GRAVY
Grand average hydropathy
- MCSG
Midwest Center for Structural Genomics
- pI
Iso-electric point
- PSI
Protein Structure Initiative
- SVM
Support vector machine
Footnotes
The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Electronic supplementary material The online version of this article (doi:10.1007/s10969-010-9080-0) contains supplementary material, which is available to authorized users.
Contributor Information
György Babnigg, Email: gbabnigg@anl.gov.
Andrzej Joachimiak, Email: andrzejj@anl.gov.
References
- 1.Gao X, et al. High-throughput limited proteolysis/mass spectrometry for protein domain elucidation. J Struct Funct Genomics. 2005;6(2–3):129–134. doi: 10.1007/s10969-005-1918-5. [DOI] [PubMed] [Google Scholar]
- 2.Koth CM, et al. Use of limited proteolysis to identify protein domains suitable for structural analysis. Methods Enzymol. 2003;368:77–84. doi: 10.1016/S0076-6879(03)68005-5. [DOI] [PubMed] [Google Scholar]
- 3.Dong A, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007;4(12):1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldschmidt L, et al. Toward rational protein crystallization: a web server for the design of crystallizable protein variants. Protein Sci. 2007;16(8):1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim Y, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5(10):853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nocek B, et al. Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J Mol Biol. 2005;354(1):91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slabinski L, et al. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics. 2007;23(24):3403–3405. doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 8.Bertone P, et al. SPINE: an integrated tracking database and data mining approach for identifying feasible targets in high-throughput structural proteomics. Nucleic Acids Res. 2001;29(13):2884–2898. doi: 10.1093/nar/29.13.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Canaves JM, et al. Protein biophysical properties that correlate with crystallization success in Thermotoga maritima: maximum clustering strategy for structural genomics. J Mol Biol. 2004;344(4):977–991. doi: 10.1016/j.jmb.2004.09.076. [DOI] [PubMed] [Google Scholar]
- 10.Goh CS, et al. SPINE 2: a system for collaborative structural proteomics within a federated database framework. Nucleic Acids Res. 2003;31(11):2833–2838. doi: 10.1093/nar/gkg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oldfield CJ, et al. Addressing the intrinsic disorder bottleneck in structural proteomics. Proteins. 2005;59(3):444–453. doi: 10.1002/prot.20446. [DOI] [PubMed] [Google Scholar]
- 12.Overton IM, Barton GJ. A normalised scale for structural genomics target ranking: the OB-Score. FEBS Lett. 2006;580(16):4005–4009. doi: 10.1016/j.febslet.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Slabinski L, et al. The challenge of protein structure determination—lessons from structural genomics. Protein Sci. 2007;16(11):2472–2482. doi: 10.1110/ps.073037907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smialowski P, et al. Will my protein crystallize? A sequence-based predictor. Proteins. 2006;62(2):343–355. doi: 10.1002/prot.20789. [DOI] [PubMed] [Google Scholar]
- 15.Price WN, II, et al. Understanding the physical properties that control protein crystallization by analysis of large-scale experimental data. Nat Biotechnol. 2009;27(1):51–57. doi: 10.1038/nbt.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 17.Marsden RL, Orengo CA. Target selection for structural genomics: an overview. Methods Mol Biol. 2008;426:3–25. doi: 10.1007/978-1-60327-058-8_1. [DOI] [PubMed] [Google Scholar]
- 18.Eddy SR. Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol. 1995;3:114–120. [PubMed] [Google Scholar]
- 19.Eddy SR. Hidden Markov models. Curr Opin Struct Biol. 1996;6(3):361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- 20.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 21.Eddy SR. What is a hidden Markov model? Nat Biotechnol. 2004;22(10):1315–1316. doi: 10.1038/nbt1004-1315. [DOI] [PubMed] [Google Scholar]
- 22.Eddy SR, Mitchison G, Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2(1):9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 23.Martelli PL, et al. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics. 2002;18(Suppl 1):S46–S53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- 24.Ward JJ, et al. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20(13):2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babnigg G, Giometti CS. GELBANK: a database of annotated two-dimensional gel electrophoresis patterns of biological systems with completed genomes. Nucleic Acids Res. 2004;32(Database issue):D582–D585. doi: 10.1093/nar/gkh089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawashima S, Ogata H, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 1999;27(1):368–369. doi: 10.1093/nar/27.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohonen T. Self-organized formation of topologically correct feature maps. Biol Cybern. 1982;43:56–69. [Google Scholar]
- 29.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25(1):8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 30.Bjellqvist B, et al. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15(3–4):529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 31.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res. 2007;35(Web Server issue):W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C, et al. Extracytoplasmic PAS-like domains are common in signal transduction proteins. J Bacteriol. 2010;192(4):1156–1159. doi: 10.1128/JB.01508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawashima S, et al. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36(Database issue):D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chothia C. Structural invariants in protein folding. Nature. 1975;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- 35.Monne M, et al. Turns in transmembrane helices: determination of the minimal length of a “helical hairpin” and derivation of a fine-grained turn propensity scale. J Mol Biol. 1999;293(4):807–814. doi: 10.1006/jmbi.1999.3183. [DOI] [PubMed] [Google Scholar]
- 36.Monne M, Hermansson M, von Heijne G. A turn propensity scale for transmembrane helices. J Mol Biol. 1999;288(1):141–145. doi: 10.1006/jmbi.1999.2657. [DOI] [PubMed] [Google Scholar]
- 37.Palau J, Argos P, Puigdomenech P. Protein secondary structure. Studies on the limits of prediction accuracy. Int J Pept Protein Res. 1982;19(4):394–401. [PubMed] [Google Scholar]
- 38.Vapnik VN. An overview of statistical learning theory. IEEE Trans Neural Netw. 1999;10(5):988–999. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- 39.Matthews BW. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta. 1975;405(2):442–451. doi: 10.1016/0005-2795(75)90109-9. [DOI] [PubMed] [Google Scholar]
- 40.Chen K, Kurgan L, Rahbari M. Prediction of protein crystallization using collocation of amino acid pairs. Biochem Biophys Res Commun. 2007;355(3):764–769. doi: 10.1016/j.bbrc.2007.02.040. [DOI] [PubMed] [Google Scholar]
- 41.Overton IM, et al. ParCrys: a Parzen window density estimation approach to protein crystallization propensity prediction. Bioinformatics. 2008;24(7):901–907. doi: 10.1093/bioinformatics/btn055. [DOI] [PubMed] [Google Scholar]
- 42.Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- 43.Munoz V, Serrano L. Intrinsic secondary structure propensities of the amino acids, using statistical phi-psi matrices: comparison with experimental scales. Proteins. 1994;20(4):301–311. doi: 10.1002/prot.340200403. [DOI] [PubMed] [Google Scholar]
- 44.Qian N, Sejnowski TJ. Predicting the secondary structure of globular proteins using neural network models. J Mol Biol. 1988;202(4):865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- 45.Richardson JS, Richardson DC. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- 46.Ponnuswamy PK, et al. Hydrophobic packing and spatial arrangement of amino acid residues in globular proteins. Biochim Biophys Acta. 1980;623(2):301–316. doi: 10.1016/0005-2795(80)90258-5. [DOI] [PubMed] [Google Scholar]
- 47.Rackovsky S, Scheraga HA. Differential geometry and polymer conformation.4. Conformational and nucleation properties of individual amino acids. Macromolecules. 1982;15(5):1340–1346. [Google Scholar]
- 48.Tanaka S, Scheraga HA. Statistical mechanical treatment of protein conformation. 5. A multistate model for specific-sequence copolymers of amino acids. Macromolecules. 1977;10(1):9–20. doi: 10.1021/ma60055a002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.