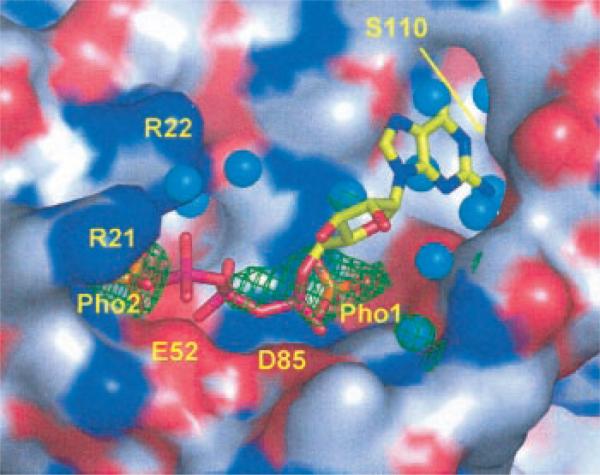
Fig. 7. Modeled GTP in the binding site of MoaB.
The surface of the protein containing the binding site is color-coded with red oxygens, blue nitrogens, and gray carbons. Experimentally observed sulfate ions (orange S and red O atoms) are shown with their corresponding 2Fo – Fc electron densities contoured at a 1σ level (green mesh). Water molecules are shown as cyan spheres. Sulfates are labeled Pho1 and Pho2 because they likely are phosphate groups in vivo. Associated with Pho1, electron density has an additional feature, which cannot be explained with the sulfate ion and which extends toward Pho2. It is likely the remnant of a phosphate-containing compound from the physiological complex. Arg21 and Arg22 are also labeled. The guanidinium group of the Arg22 side chain is not entirely visible in our structure but is added in this figure to show its proximal position and to avoid the creation of an apparent extended surface depression that could lead to erroneous interpretations. The ridge, separating two deeper cavities on either side of Pho1, is formed by two conserved motifs, Gly77-Gly78-Thr79-Gly80 and Pro137-Gly138-Ser139. Modeled GTP is shown with stick model color coded as yellow carbons, blue nitrogens, red oxygens, and magenta phosphors. The locations of the Glu52 and Asp85 carboxyl groups are indicated with labels, and the location of Ser110 is shown with an arrow.