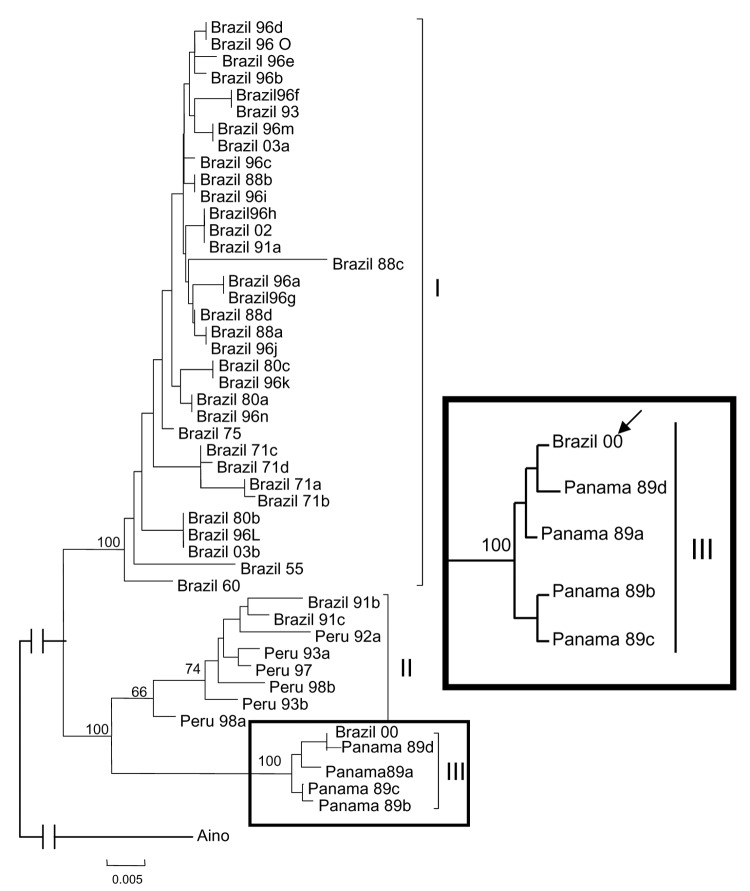
Figure 2.
Phylogeny of Oropouche virus (OROV) strains isolated from different sources and periods by using the neighbor-joining and maximum parsimony methods. Bootstrap values were assigned over each internal branch nodes, and highest values were indicated by continuous arrows showing the presence of at least 3 lineages or genotypes (I, II, and III) of OROV. Bootstrap values for the 3 representative genotype clades are placed over each respective branch node. The black arrow indicates the position of the strain BeAn 626990 (Brazil 00) in the tree. The Aino N gene nucleotide sequence was used as an outgroup to root the tree. The scale bar represents 5% nucleotide sequence divergence.