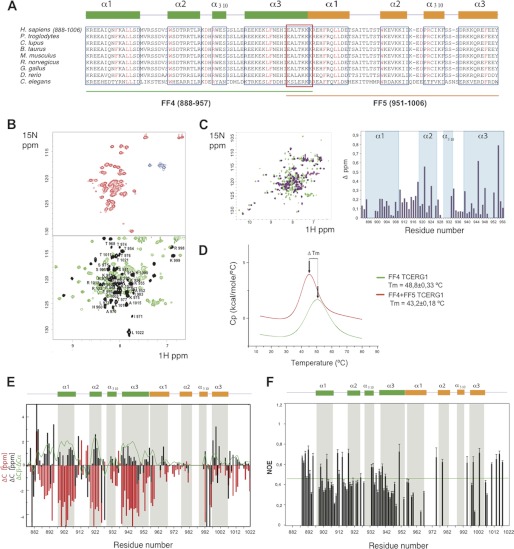
FIGURE 2.
15N-HSQC NMR spectra and thermal unfolding of FF4-FF5. A, amino acid alignment of TCERG1 homologues. Secondary structure elements of FF4 and FF5 are drawn above the sequences in green and orange, respectively. Boxed sequences in blue are the residues that form the α-helices. The overlapped residues between FF4 and FF5 are shown in a red box. Strictly conserved residues are highlighted in red. B, 15N-HSQC spectra at 298 K for FF5 and FF4-FF5 constructs. The NMR data show the improvement of FF5 backbone amino acid dispersion (shown in black) observed in the pair when compared with that of the isolated construct. C, overlapped 15N-HSQC spectra at 298 K for FF4 and FF4-FF5 constructs and chemical shift differences bar representation of FF4 residues. D, thermal unfolding of the FF4 and FF4-FF5 domains as monitored by DSC. The difference in Tm is shown as an arrow connecting both maxima. E, secondary structure elements of the FF4-FF5 pair. Chemical shift distribution of Cα and Cβ. The ratio of Cα and Cβ is shown as a green line. The majorities of the values are positive, indicating the presence of a helical structure. F, heteronuclear {1H}-15N NOE. Unassigned residues, proline residues that lack a proton amide, and overlapped peaks were excluded from the analysis. Heteronuclear NOE values show that the pair of FF4-FF5 has secondary structure throughout the construct.