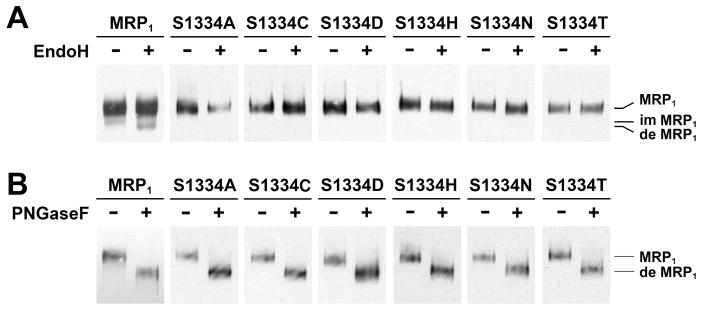
Figure 1.
Substitution of S1334 in the Walker A motif of NBD2 with an amino acid that eliminates the hydroxyl group at this position has no effect on protein folding. (A) All of the MRP1 mutants mainly form complex-glycosylated mature proteins at 37 °C. Cells grown at 37 °C were lysed with 2% SDS and incubated in the absence (−) or presence (+) of endoglycosidase H according to the method described in Experimental Procedures. (B) The complex-glycosylated MRP1 proteins are sensitive to PNGase F. The same cell lysates in panel A were incubated in the absence (−) or presence (+) of PNGase F according to the method described in Experimental Procedures. MRP1, im MRP1, and de MRP1 on the right indicate the complex-glycosylated mature MRP1, the core-glycosylated immature MRP1, and the deglycosylated MRP1 proteins that were detected in Western blots by employing the monoclonal antibodies 42.4 and 897.2 against MRP1 (8).