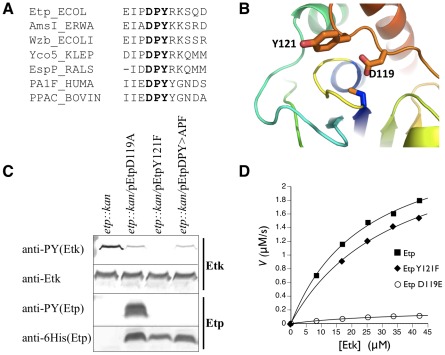
Figure 3. Identification of the phosphorylated Etp residue.
(A) The region containing the DPY motif (in bold) of Etp is compared with the corresponding region of several members of the LMW-PTP family. They include AmsI of Erwinia amylovora, the E. coli-encoded Etp paralog Wzb, Yco5 of Klebsiella pneumoniae, EspP of Ralstonia solanacearum, the human PA1F protein, and the bovine PPAC protein encoded by the ACP1 gene. Numbering is according to the Etp sequence. (B) Homology model of the Etp structure in the absence of bound substrate based on the NMR structure of Wzb [19]. Side chains Tyr121 and Asp119 are shown in sticks. The phosphate binding loop (yellow) and catalytic cysteine C13 are in the center. (C) EPEC mutant Δetp::kan was transformed with plasmids expressing different Etp mutants all of which had C-terminal 6His tags. Proteins were extracted from the different cultures and were subjected to Western blot analysis with anti-Etk, anti-6His and anti-PY antibodies as indicated. The corresponding strain is indicated above each of the lanes. (D) In vitro kinetics of Etp and variants with phosphorylated MBP-Etk as substrate. The graph plots the rate of inorganic phosphate produced versus substrate concentration for Etp, EtpY121F, and EtpD119E. Kinetic constants are in Table 1.