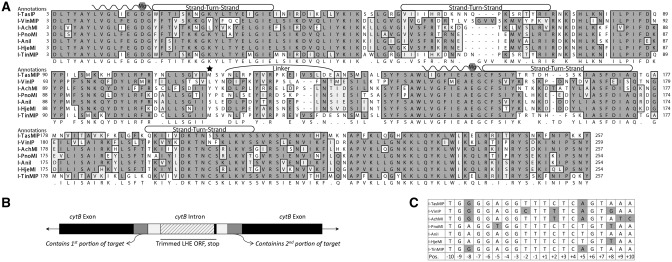
Figure 1.
Predicted homologs and targets. (A) The alignment of the I-AniI homologs with the residues shaded by chemical similarity. LAGLIDADG motifs are marked by waved lines. Conserved Mg++-coordinating residues and DNA-contact rich strand-turn-strand regions are also annotated. The homologous serine 111, a residue important for increased catalytic activity in I-AniI, is starred. The map was generated by MacVector using Gonnet-weighted pairwise and multiple sequence alignments with residue-specific and hydrophilic penalties. Residue numbering was matched to I-AniI, based on the first LAGLIDADG motif. (B) Schematic depicting the original host gene (black) with intron insertion (white) from which the LHE ORF sequences were taken and the exon/intron junctions used to predict target sequences (gray). (C) Predicted targets for each homolog, derived by comparing flanking intron/exon regions for each intronic LHE with those from I-AniI; differences therefrom are shaded.