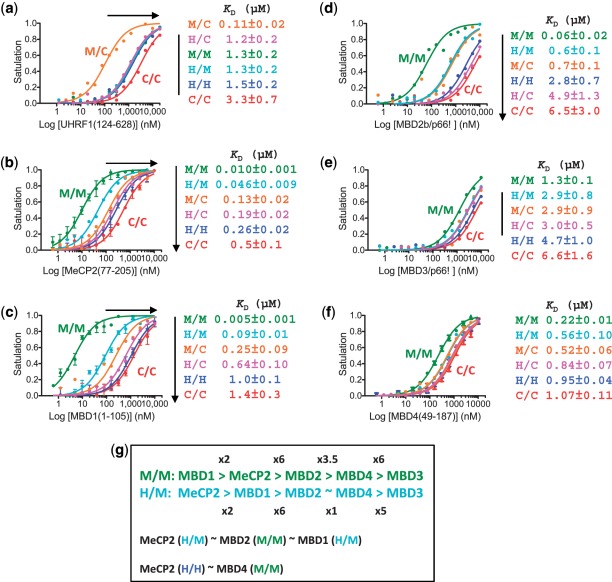
Figure 2.
Effect of hydroxymethylation on DNA methylation ‘readers’. Binding affinities of UHRF1 (residues 124–628) (a) and five MBD proteins (b–f) for a double stranded oligonucleotide containing a single CpG site with one of the six different modification states: unmodified (C/C), fully modified (M/M, H/H), hemi-(hydroxy)methylated (M/C, H/C) or hemi-methylated/hemi-hydroxymethylated (H/M) CpG site. Binding was assessed by fluorescence polarization. (a) UHRF1 has a strong preference for binding the hemi-methylated CpG (M/C) site. (b and c) MeCP2 and MBD1 have the strongest binding to fully methylated CpG (M/M). Although there is significantly lower affinity among the MBDs for H/M CpG dinucleotides (5-fold decreased affinity for MeCP2 and 18-fold for MBD1), this substrate is still preferred over hemi-methylated DNA (M/C), and is bound by MeCP2 and MBD1 with an affinity similar in magnitude to MBD2 (d), and greater affinity than MBD3 (e) and MBD4 (f) bind to fully methylated DNA (M/M). (g) Summary of relative binding affinities (by factor of x) of five MBD proteins for M/M and H/M substrates (top two lines). Note that the binding affinities of MeCP2 (and MBD1) for H/M and H/H are in the same order of magnitude as that of MBD2 and MBD4 to M/M substrate, respectively (lines 3 and 4).