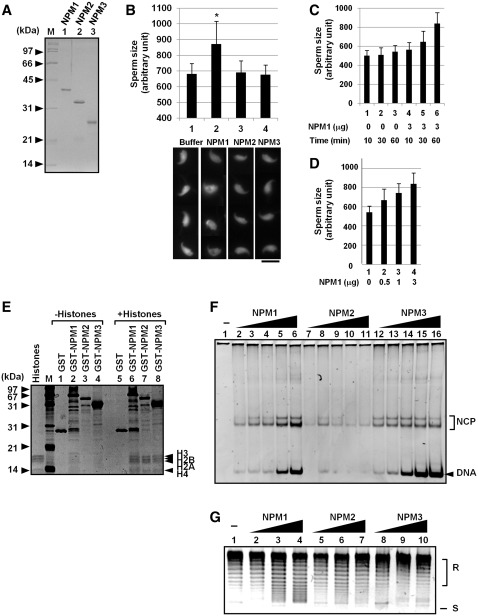
Figure 3.
Sperm chromatin remodeling by human NPM proteins. (A) Purified proteins. His-tagged NPM proteins were expressed in E. coli and purified. Proteins (200 ng each) were separated by 12.5% SDS–PAGE and visualized with CBB staining. Positions of molecular weight markers are indicated at the left side of the panel. (B) Sperm chromatin decondensation by NPMs. Mouse sperm nuclei (1 × 105) were incubated for 60 min without or with 3 µg of purified NPM1, NPM2, or NPM3. Sperm nuclei were fixed with formardehyde and stained with DAPI. Bar at the bottom of the panels indicates 10 µm. Four typical sperm nuclei are shown for each sample. Size of the sperm nuclei (n > 30) were estimated by Image J software and averaged. Results are means ± SD and statistical P-values were calculated and indicated * for P < 0.05. (C and D) Sperm chromatin decondensation by NPM1. Sperm nuclei were incubated in the presence or absence of NPM1 and incubated for 10–60 min as indicated at the bottom of the graph (C) or for 60 min (D). Size of sperm nuclei was analyzed as in (B). (E) Histone-binding activity of human NPM proteins. GST, GST-tagged NPM1, NPM2, and NPM3 (1 µg each) were mixed without (lanes 1–4) or with core histones (1 µg, lanes 5–8) purified from HeLa cells in the buffer containing 150 mM NaCl. The mixtures were recovered by glutathione sepharose beads. The proteins were separated by 15% SDS–PAGE and visualized with silver staining. Positions of molecular weight markers and core histones are shown at the left and right side of the panel, respectively. (F) Histone transfer assay. Increasing amounts of His-tagged NPM1, NPM2 and NPM3 (lanes 2–6, 7–11 and 12–16, respectively) (50, 100, 200, 300 and 400 ng for each protein) were incubated with core histones (100 ng) and incubated. Then the 147-bp DNA fragment (30 ng) were added and further incubated at 37°C for 30 min. The mixtures were separated by 6% PAGE in 0.5× TBE. DNA was visualized by staining with GelRed. Positions of free DNA and nucleosome core particle (NCP) are indicated at the right side of the panel. (G) Supercoiling assay. His-tagged NPM1, NPM2 and NPM3 (100, 300, and 1000 ng for lanes 2–4, 5–7 and 8–10) were incubated with core histones (100 ng). Then plasmid DNA (100 ng) treated with topoisomerase I was added to NPM-histone mixture and incubated. Plasmid DNA was purified and analyzed by 1% agarose gel electrophoresis in 1× TBE and visualized with GelRed staining. Positions of relaxed (R) and supercoiled (S) DNA are indicated at the right side of the panel.