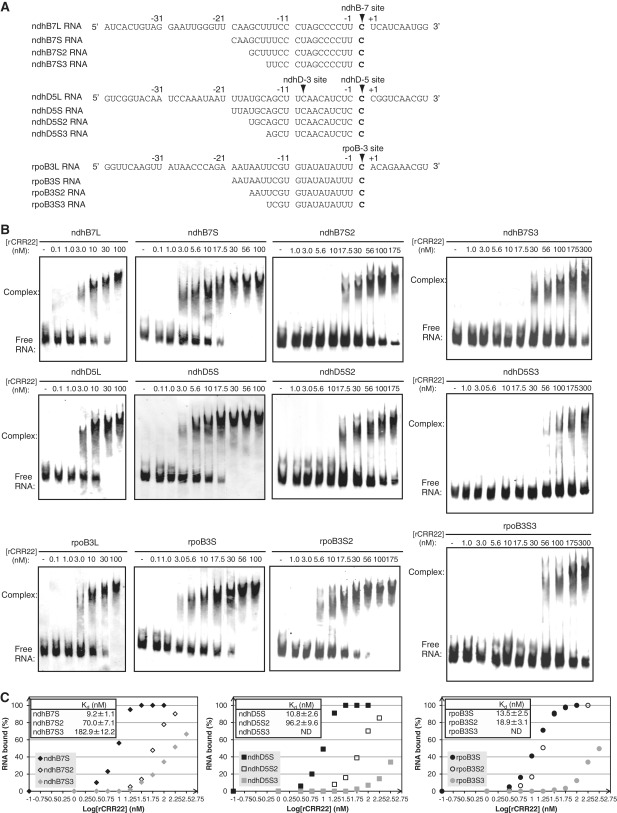
Figure 5.
GMS assays with rCRR22 and target RNAs. (A) RNA sequences used as probes are shown. Editing sites of ndhB-7, ndhD-5 and rpoB-3 are indicated in bold and are marked with arrowheads. (B) GMS assays were performed with the indicated concentrations of rCRR22 and labeled RNAs (ndhB7L, ndhB7S-S3, ndhD5L, ndhD5S-S3, rpoB3L and rpoB3S-S3), as described in the ‘Materials and Methods’ section. (C) Equilibrium Kd of rCRR22 for the ndhB7S-S3 (left), ndhD5S-S3 (middle) and rpoB3S-S3 (right) probes. rCRR22 concentrations and fractions of RNA bound in each lane are plotted. The Kd calculation assumes a 1:1 interaction between the RNA and the protein. The Kd values and each data point are means ± SD of three experiments performed with the same rCRR22 preparation. All of the GMS assays were performed with the same preparation of rCRR22 and within 2 weeks after purification.