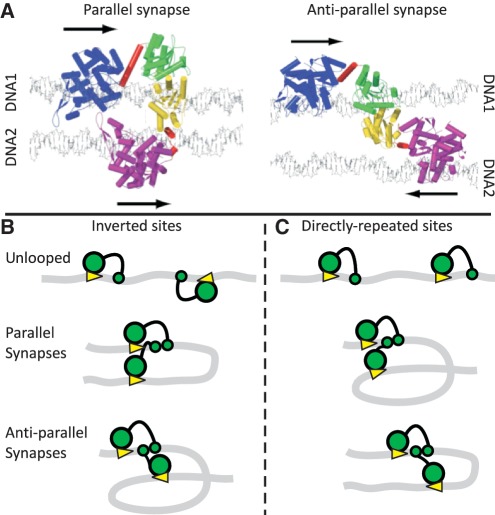
Figure 1.
Synaptic complexes and loop topologies. (A) Shown here (with permission from Elsevier Press) are the models of the FokI synapse from Vanamee et al. (31). The complexes feature two monomers of the FokI endonuclease holding together two DNA duplexes (1 and 2) in either parallel or anti-parallel arrangements, with the catalytic domains from the two monomers each positioned to cut one strand of DNA1: the arrows denote the 5′→3′ direction of the recognition sequences (5′-GGATG-3′). The DNA recognition and catalytic domains of the protein bound to DNA1 are in blue and green, respectively, while the equivalent domains of the protein bound to DNA2 are in purple and yellow: α-helical segments are marked as cylinders. In both monomers, the linker region is in red. Note that in the model of the parallel synapse, the linker region of the protein bound to DNA1 contains a long α-helix, which is snapped into two for the protein bound to DNA2. (B and C) The DNA molecules (grey ribbons) carry two recognition sites for FokI (yellow arrowheads, pointing towards the downstream sites for DNA cleavage), in either inverted (IF) or directly repeated (DF) orientations along the DNA. The recognition and catalytic domains of the FokI monomers bound to each site are shown as large and small green circles, respectively. (B) On an IF DNA, juxtaposition of the sites in the parallel manner must bend the intervening DNA through 180° while juxtaposition in the anti-parallel alignment requires it to be bent through 360°. (C) The converse applies to a DF DNA: the parallel synapse traps a convoluted 360° loop and the anti-parallel a simple 180° loop.