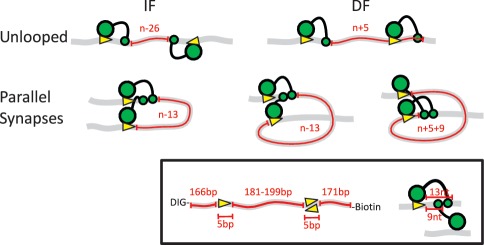
Figure 1.
Schematic representation of the possible orientations of two recognition sites for FokI in cis and the resulting loop topologies. The FokI restriction endonuclease recognizes a 5-bp non-palindromic sequence and cuts 9 and 13-nt downstream of the site, in top and bottom strands, respectively (insert). The DNA molecules are shown as grey ribbons with two recognition sites for FokI (yellow arrowheads) in cis, in either inverted (IF) or directly repeated (DF) orientations (as indicated by the direction of the arrowheads): ‘n’ is the absolute distance in base pairs between the recognition sites, measured from the base pairs immediately following the upstream recognition site to that immediately before the downstream. The FokI monomer is shown as a large green circle (the DNA recognition domain bound to its target sequence) connected to a small green circle (the catalytic domain that before dimerization binds to the scissile bond 13 nt away). Hence, in the unlooped state, the length of DNA between the two catalytic domains (noted in red) is n − 26 bp in the IF construct and n + 5 bp in the DF construct: these lengths denote the length of DNA involved in the loop capture process (i.e. the ‘loop size’ for capture). When the catalytic domains of two monomers on the DNA associate to the dimer, the sites become aligned in a parallel arrangement, giving rise to two different loop topologies: a DNA loop with a 180° bend from the IF DNA (left), or a more extended loop with a 360° bend from the DF DNA (right: note that the latter has two alternative configurations, depending on whether the dimer is assembled at the upstream or the downstream recognition site). In each case, the number of base pairs trapped between the two protein monomers (noted in red) differs from the absolute separation of the sites (n) as shown: the lengths of the trapped segments correspond to ‘loop size’ for the release process.