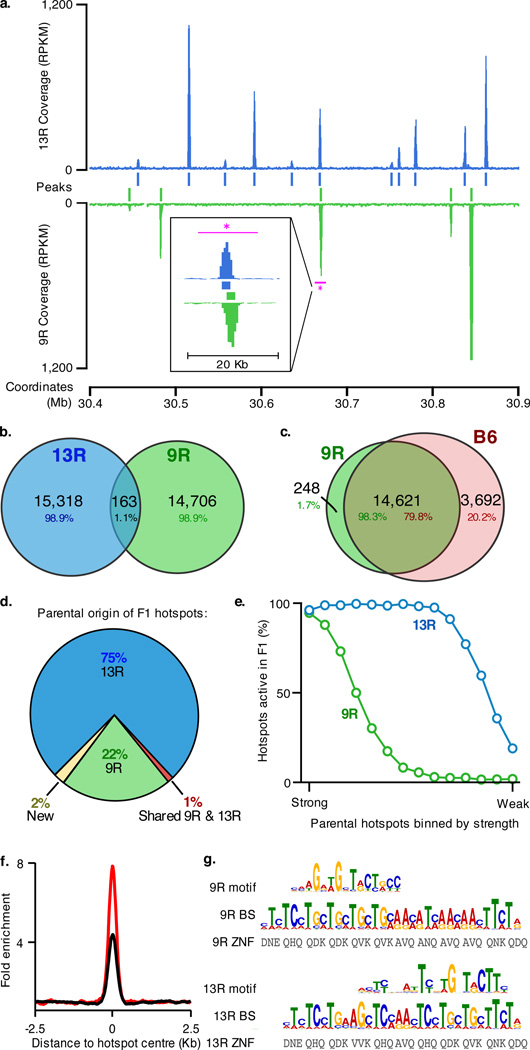
Figure 1.
DSB hotspots localize to different loci in 9R and 13R mice. a. DSB hotspots in a representative 0.5 Mb region on chromosome 3. The magenta bar indicates the region shown in the insert. This illustrates two adjacent, yet distinct hotspots in 9R and 13R. Data are smoothed using a 1 Kb sliding window (step 100 bp). Coverage is given as reads per Kb per million (RPKM) b. Overlap between 9R and 13R hotspots. Here and in subsequent panels, only overlaps in the central 400 bp of hotspots are counted. c. Overlap between 9R and C57Bl/6 (B6) hotspots. The excess of B6 hotspots is due to higher ChIP enrichment in this ChIP-Seq sample. B6 hotspots not found in 9R are a weak subset. d. Parental origin of DSB hotspots in the F1 (9R × 13R) mice. e. Stronger 9R and 13R hotspots are active in F1 progeny. The top 14,000 hotspots for each background were binned by strength and the overlap with F1 hotspots assessed for each bin. f. Distribution of hits to the 9R motif (black) and 13R motif (red) around their respective hotspots. Data are plotted in 200 nt steps. g. Alignment of 9R and 13R motifs to the predicted PRDM9 binding sites (see methods). ZNF, Zn finger contact residues; BS, predicted PRDM9 binding site.