Abstract
We have developed a chemical reagent that recognizes all naturally occurring DNA bases, a so called universal reader, for DNA sequencing by recognition tunnelling in nanopores.[1] The primary requirements for this type of molecules are the ability to form non-covalent complexes with individual DNA bases and to generate recognizable electronic signatures under an electrical bias. 1-H-imidazole-2-carboxamide was designed as such a recognition moiety to interact with the DNA bases through hydrogen bonding. In the present study, we first furnished a synthetic route to 1-H-imidazole-2-carboxamide containing a short ω-functionalized alkyl chain at its 4(5) position for its attachment to metal and carbon electrodes. The acid dissociation constants of the imidazole-2-carboxamide were then determined by UV spectroscopy. The data show that the 1-H-imidazole-2-carboxamide exists in a neutral form between pH 6–10. Density functional theory (DFT) and NMR studies indicate that the imidazole ring exists in prototropic tautomers. We propose an intramolecular mechanism for tautomerization of 1-H-imidazole-2-carboxamide. In addition, the imidazole-2-carboxamide can self-associate to form hydrogen bonded dimers. NMR titration found that naturally occurring nucleosides interacted with 1-H-imidazole-2-carboxamide through hydrogen bonding in a tendency of dG>dC≫dT> dA. These studies are indispensable to assisting us in understanding the molecular recognition that takes place in the nanopore where routinely used analytical tools such as NMR and FTIR cannot be conveniently applied.
Keywords: DNA, hydrogen bonds, recognition tunneling, self association, universal reader
Introduction
DNA sequencing is a pivotal tool in rapidly expanding personal genomics where individual genomes are genotyped and bioinformatically analyzed to find disease association of genes and loci. The sequencing of DNA and RNA will eventually form the basis of personalized medicine.[2] In spite of its monumental accomplishments culminating in the completion of Human Genome Project, the classical Sanger method[3] has limited use where a large number of genome sequences are involved due to its high cost and low throughput. Current advances in next generation sequencing (NGS) have brought the whole genome cost down to thousands of dollars and the sequencing timescale down to a number of days. There are several different types of NGS DNA sequencers available on the market,[4] all of which are based on enzymatic reactions and are dominated by optical detection. DNA segments (or “reads”) sequenced by these NGS technologies vary in length from 35 to <1000 base pairs. Short reads hamper de novo assembly of complex genomes from scratch, suffering from duplications and repeats, as well as the fragmentation of genomic sequences.[5] As a result, data from NGS are often assembled using published human genomes as references, an approach that may create distorted personal genomes.[6] To overcome these caveats of NGS, nanopores with diameters comparable to that of a DNA double helix (ca. 2 nanometers) have been investigated as a physical means for the sequencing.[7] A nanopore should be able to read very long segments, meanwhile simplifying sample preparation and eliminating the use of costly biochemical reagents, such as polymerases or ligases. In principle, a single stranded DNA, while passing through a nanopore, blockades ionic current and generates electronic signals corresponding to its base compositions and the order of bases in the sequence. It has been demonstrated that the ion current blockades can used to distinguish all four nucleotides and 5-methylcytidine monophosphate (5-methyl C) if they are individually passed into the pore,[8] and single bases in a DNA oligomer can be read if trapped in an MspA pore at a double strand-single strand junction.[9] Nevertheless, nanopores to date have not shown the capability of reading a DNA sequence. One of the reasons is that the current blockade is the sum of effects from all the DNA bases inside the nanopore,[10] complicating a single base resolution.
Measuring the electron tunneling current that is created transversely across a translocating DNA molecule has been proposed as an alternative to the measurement of ion current blockades.[7a,11] We have recently demonstrated by means of a Scanning Tunneling Microscope (STM) setup that a DNA base flanked by other bases in a short DNA oligomer can be identified in a nanogap consisting of two gold electrodes functionalized with mercaptobenzamide in aqueous solution.[12] The benzamide specifically functions as a recognition moiety, which presumably interacts with DNA bases through hydrogen bonding. We found that the benzamide can read all DNA bases, A, C, mC, G, except T. In addition, there are significant overlaps of electric signal peaks among A, mC, G, which would compromise the base-calling accuracy. Although the selectivity of benzamide can be improved by optimizing the experimental conditions and including other characteristics of the tunneling noise, a highly discriminating universal reader molecule will ultimately fulfill a better resolution and simplified data analysis.
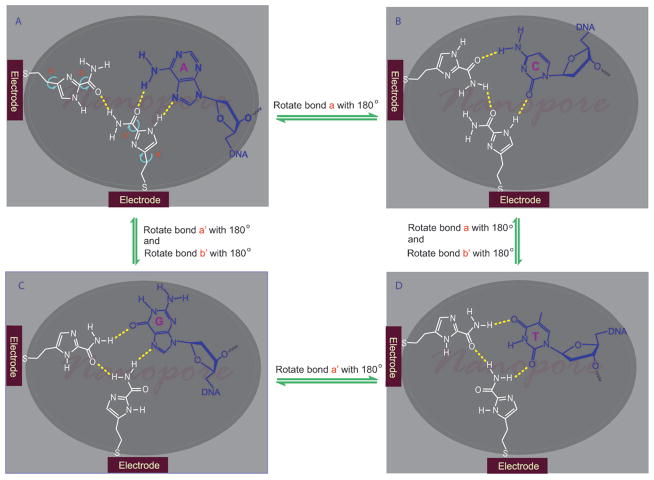
In one configuration of the nanopore sequencing devices under development, atomically thin electrodes are embedded in the dielectric film with their ends exposed to the nanopore.[13] We envision that as a single stranded DNA molecule translocates through a nanopore, the inside water molecules are squeezed out of the nanopore, rendering the interior of the nanopore less hydrophilic in comparison to its outside. Thus, unlike in the aqueous solution, the hydrogen bonding interactions should be conceivable inside the nanopore. Based on this hypothesis, we have drafted a set of general guidelines for design of a univesal reader. The molecule should contain multiple hydrogen bonding sites with well balanced numbers of donors and acceptors, have high conductance, and be small enough in size to sit in a nanopore without impeding the DNA translocation. In addition, it should have variable conformations in order for one to fit all. The universal reader must also be electrochemically stable under physiological conditions. Our first choice was five-membered aromatic nitrogen heterocycles because of their π-excessive (6 π electrons shared by 5 atoms) nature[14] and built-in hydrogen bonding sites. Bergstrom and coworkers have synthesized 4(5)-(2′-deoxy-β-D-ribofuranosyl)-1-H-imidazole-2-carboxamide as a degenerate DNA base analog.[15] It was unclear how this molecule interacts with DNA bases since its base pairing properties have not been examined. At first glance, 1-H-imidazole-2-carboxamide is an aromatic molecule that contains several hydrogen bonding sites. However, when the imidazole-2-carboxamide moiety is attached to an electrode through a flexible linker, it can freely rotate and its amide group can rotate as well, as illustrated in Figure 1. In a nanopore with a diameter of 2 nm, an imidazole-2-carboxamide molecule in one electrode can reach the same molecule in another electrode to form a molecular bridge through hydrogen bonding for the electron tunneling. Due to the changeable conformations of the imidazole-2-carboxamide molecule, the bridge can exist in multifaceted configurations, each of which should selectively recognize one of DNA bases and form an electrically distinguishable triplet structure (Figure 1A, B, C, D). With two free rotating bonds, two imidazole-2-carboxamides can statistically form as many as 16 different facets. Thus, it should be possible for such a single molecule to read all the DNA bases and their modifications, such as 5-methylcytosine and N6-methyladenine which have different π-electron distributions from their parent bases. In the present work, we have developed an effective way to synthesize 4(5)-substituted-1-H-imidazole-2-carboamides and studied physicochemical and hydrogen bonding properties of the imidazole-2-carboxamide moiety in solution. The goal was to chemically understand the imidazole molecule, laying down a basis for studying its interactions with DNA bases and electron tunneling in the nanopore.
Figure 1.
An illustration of 1-H-imidazole-2-carboxamide as a universal reader, which reads each of DNA bases by changing its conformations to form a different hydrogen bonding pattern (in an actual device fabricated by E-beam lithography and atomic layer deposition (ALD) processes, the atomically thin electrodes are embedded in a dielectric film with their ends exposed to the nanopore).
Results and Discussion
Synthesis of 1-H-imidazole-2-carboxamide containing a short linker at its 4(5) position
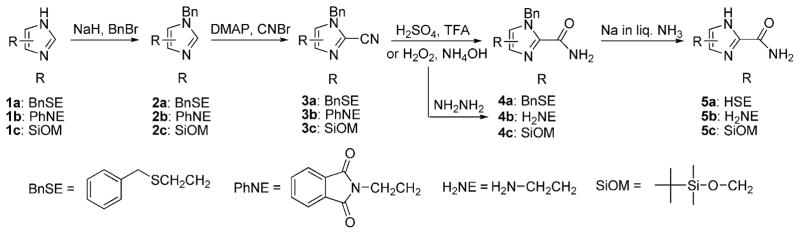
We have synthesized a number of 1-H-imidazole-2-carboxamides composed of a short ω-functionalized alkyl chain at their 4(5) positions for attachment to metal electrodes. In general, the 1-H-imidazole derivatives exist in tautomeric forms that are chromatographically inseparable. No attempt was made to separate the tautomers during the synthesis. Since a variety of 4(5)-alkylated imidazoles were commercially available or reported in literature, the main efforts of the synthesis was finding a facile method to introduce an amide group to the 2 position of the imidazole ring. As illustrated in Scheme 1, 4(5)-(2-thioethyl)-1-H-imidazole-2-carboxamide (5a) and 4(5)-(2-aminoethyl)-1-H-imidazole-2-carboxamide (5b) were synthesized from 4(5)-(2-(benzylthio)ethyl)-1-H-imidazole (1a) and N-phthaloyl histamine (1b), respectively. The thiol and amine function as anchor groups for attaching the molecule to either metal or carbon electrodes. In the same way, 4(5)-(tert-butyldimethylsilyloxymethyl)-1-H-imidazole-2-carboxamide (5c) was synthesized from 4(5)-(tert-butyldimethylsilyloxymethyl)-1-H-imidazole (1c), and used as a model compound for NMR studies on hydrogen bonding properties of 1-H-imidazole-2-carboxamide in organic solvents (vide infra). We have explored two routes for synthesizing these 1-H-imidazole-2-carboxamide compounds mentioned above. The 2 position of imidazole can be substituted with a formate ester[16] or a cyano group,[17] both of which can readily be converted into an amide group. We found that the cyano route gave us a better yield overall. First, compound 1a, 1b, and 1c were converted into the 1-H nitrogen protected products (2a, 2b, 2c) by reacting their sodium salts (generated in situ by treating them with NaH) with benzyl bromide. NMR confirms that each of them is a mixture of two isomers with the benzyl group at N1 and N3. We have tried different protecting groups such as trityl and tert-butyloxycarbonyl (Boc), finding that benzyl was the most effective protecting group for the following cyanating reaction. The cyano group was introduced into the 2 position of the imidazole ring of 2a, 2b, and 2c by treating them with 1-cyano-4-(dimethylamino)pyridinium bromide (CAP). CAP was generated in situ by mixing equivalent amounts of cyanogen bromide and 4-(dimethylamino)pyridine in dimethyl formamide (DMF) at 0°C. A 2.5 fold of CAP gave the best yield. The cyano group of 3a was converted into an amide (4a) by hydrolysis in a mixture of sulfuric acid (20% by volume) and trifluoroacetic acid (18% by volume) in a fair yield (46%).[18] Under the basic condition and in the presence of hydrogen peroxide,[19] the hydrolysis could give us 4b from 3b (the phthaloyl group was completely removed by hydrazine) and 4c from 3c, but failed to furnish the desired product 4a from 3a because the benzylthio group was oxidized and eliminated from 3a. Final products 5a, 5b, and 5c were obtained by removing the benzyl groups with sodium in liquid ammonia. The tert-butyldimethylsilyloxymethyl group survived from the debenzylation conditions. 5c was separated in a fair yield (35%).
Scheme 1.
Synthetic route to 4(5)-substituted-1-H-imidazole-2-carboxamides. BnBr=benzyl bromide; DMAP=4-(dimethylamino)pyridine; TFA=trifluoroacetic acid.
Acid dissociation constants (pKa) of 1-H-imidazole-2-carboxamides
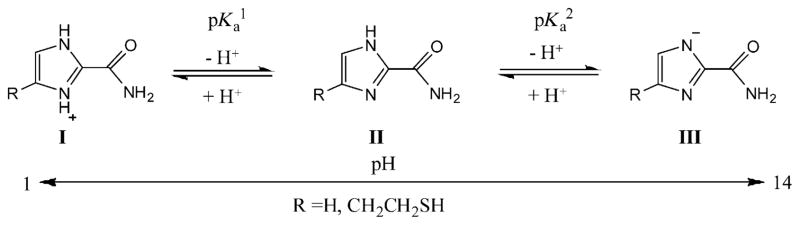
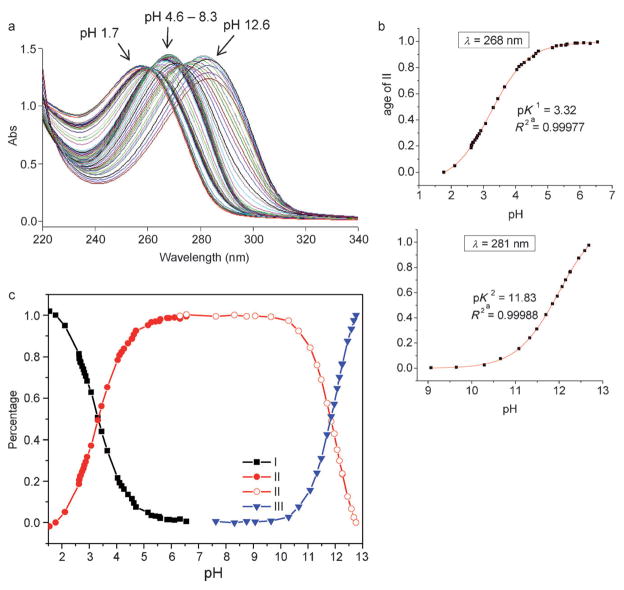
Imidazole is amphoteric, which can either gain a proton via its imine nitrogen N3 (as a base) to form an imidazolium, or lose its NH proton (as an acid) to form an imidazolide in aqueous solution, as shown in Scheme 2.[20] The introduction of an amide group to the imidazole should alter both its acidity and basicity. We determined acid dissociation constants of 1-H-imidazole-2-carboxamide in buffered solution by means of UV spectroscopy (Figure S1 in the Supporting Information), giving pKa1=3.29 and pKa2 between 12 and 13. As expected, they are smaller than those of the parent imidazole molecule (pKa1=7.0, pKa2=14). To examine effects of 4(5) substitution on acidity and basicity of the imidazole, we first determined pKas of 5a in aqueous solutions (not buffered). The UV spectra of 5a was recorded at different pHs ranging from 1 to 13 (Figure 2a). The measured pKas in this pH range should directly be related to protons on the nitrogen atoms of the imidazole ring. In general, an amide proton has its p Ka value below 0 as a base and above 15 as an acid.[21] Our NMR data show that the NH proton (δ=10.1 ppm) of the imidazole-2-carboxamide appears at the downfield of its amide protons (δ=6.2 ppm) in chloroform (see below). That indicates that the NH proton is more acidic and should be deprotonated ahead of the amide in the process of the titration with base. From Figure 2a, we observe hypochromic shifts of the λmax from 258 to 268 to 281 nm with increasing pH and two isosbestic points respectively at 261 and 274 nm, indicating that there are two UV absorbents existing in each of the equilibriums. The degree of dissociation versus pH curves were well fit to a sigmoidal function (R2>0.99), which yielded pKa1=3.32 and pKa2= 11.83 for 5a (Figure 2b). We also determined the pKas of 5a in buffered aqueous solutions with the result that pKa1= 3.33 and pKa2 between 12 and 13 (Figure S2 in the Supporting Information). These studies show that the 4(5) substitution has no noticeable effects on pKa values of 1-H-imidazole-2-carboxamide. However, the pKa2 value of 5a in the non-buffered solution is smaller than in the buffered solution. This indicates that ions may impede release of the NH proton from the imidazole ring. Figure 2c shows the distribution of the three forms of the 1-H-imidazole-2-carboxamide versus pH. Between pH 6 to 10, the imdazole-2-carboxamide exists solely in a neutral form, which is important for tunneling experiments carried out in physiological conditions (pH ca. 7). It should be noted that the thiol group in 5a does not interfere the pKa measurement significantly. Thiol (RSH) is a weak acid with pKa about 10 and forms thiolate (RS−) under basic conditions. Although the thiolate has strong UV absorbance at 238 nm, the UV absorbance of thiol above 250 nm and thiolate above 275 nm is negligible.[22]
Scheme 2.
Protonation and deprotonation equilibria of imidazole-2-carboxamide in aqueous solution.
Figure 2.
a) UV spectra of 5a at different pHs ranging from 1.7 to 12.6. b) Degree of dissociation/pH profile of 5a at 268 and 281 nm. c) Distribution of the three forms of the imidazole with pH, calculated based on UV absorbance at 281 nm, 268 nm, and 258 nm from Figure 2a, respectively (see the Experimental Section for details).
Structure, tautomerism, and self association of 1-H-imidazole-2-carboxamide
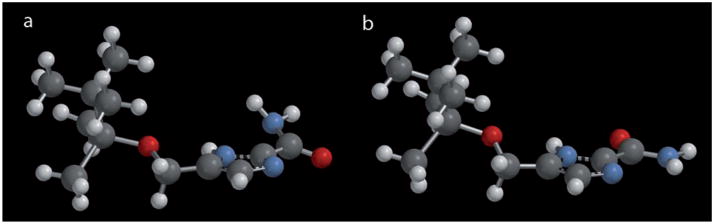
Owing to its suitable solubility in organic solvents, 5c was employed as a model compound to study the hydrogen bonding interactions of imidazole-2-carboxamide. We first studied its molecular structure by DFT. Since the amide group is connected to the imidazole through a σ bond, it can rotate freely and exist in a conformation with its NH2 either cis or trans to the NH of the imidazole ring (Figure 3). The DFT calculation indicates that the amide would primarily take the trans conformation (Figure 3b). As shown in Figure 3a, in the cis conformation, the NH2 group is pushed out of the imidazole plane by the NH group, losing conjugation with the imidazole ring. There is a difference of approximately 10.5 kcalmol−1e in total energy between the cis and trans conformations (Table S1 in the Supporting Information).
Figure 3.
DFT model of 5c with a) N–H of the imidazole cis with respect to the NH2 group of the carboxamide. b) N–H of the imidazole trans with respect to the NH2 group of the carboxamide.
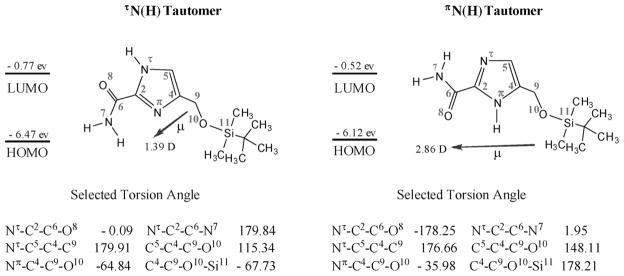
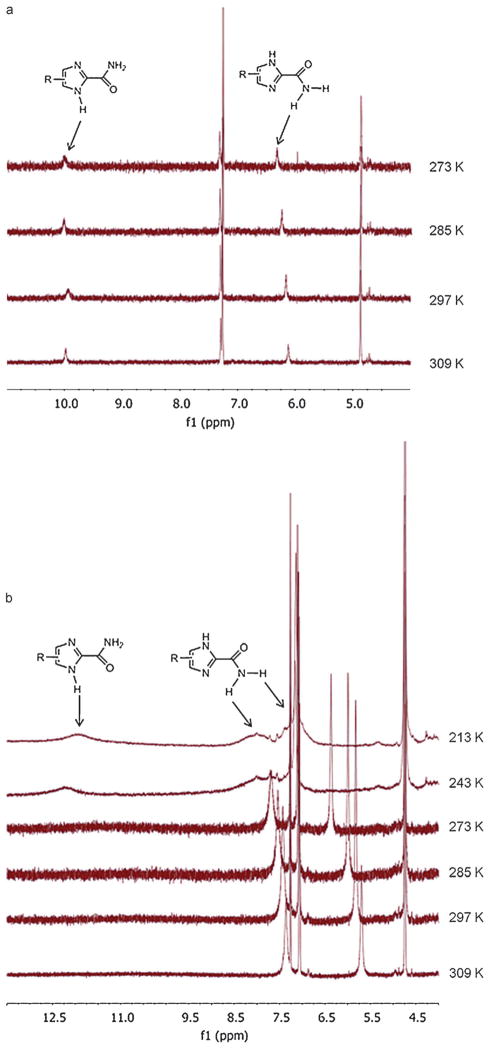
Another important feature of the 1-H-imidazole-2-carboxamide molecule is its tautomeric proton (NH) that sways between two nitrogen atoms of the imidazole ring. We studied the molecular structures of two prototropic tautomers of 5c with the amide situated at its trans conformation in detail by DFT calculations using B3LYP in combination with 6-311+ +G** basis sets in vacuum, followed by B3LYP/6-31G* to calculate their NMR chemical shifts in chloroform. Some of characteristic structure parameters are summarized in Figure 4. In the DFT model, the SiOM group stays on the top of the imidazole amide plane, projected in the direction opposite to the amide and away from the imidazole ring (see Figure 3) so that the SiOM group would not interfere in hydrogen bonding interactions related to the amide group. Furthermore, the NH proton does not form intramolecular hydrogen bonds with either the SiOM or the amide group, referring to the geometric definition for hydrogen bonds: d≤2.35 & and 100° ≤α≤180° (d=length of the hydrogen bond, α=angle of the hydrogen bond).[23] However, the SiOM group has some weak interactions most likely through its oxygen with the NH in the πN(H) tautomer. In the DFT structure of πN(H) tautomer, the torsion angle Nπ-C4-C9-O10 is −35.98°, much smaller than the same one (−64.84°) in the τN(H) tautomer. This places the oxygen near the NH with the O to H distance about 2.6 & in the πN(H) tautomer, which deshields the proton and moving its chemical shift downfield a bit in comparison to the shift in the NH in the τN(H) tautomer in NMR spectroscopy (see Scheme 3). The DFT calculation shows that the πN(H) tautomer is slightly more stable than the τN(H) tautomer by −1.08 kcalmol−1 in the total energy (Table S1 in the Supporting Information), which is so small that we conclude that both tautomers should concomitantly exist in solution without a marked preference. In a 2D NOSEY proton NMR spectrum of 5c (15 mM in chloroform), we observe the NOE cross peaks of the NH correlating with C5H, C9H, and CH3 protons, and no cross peaks from correlation of the NH with the amide NH2 (Figure S3 in the Supporting Information). These results can be interpreted by the presence of the tautomers and the amide in its trans conformation. 5c was also studied by variable temperature (VT) NMR. In a low concentration (2 mM), both NH and amide protons appear as single peaks from 273 to 309 K (Figure 5a), indicating that the tautomeric proton are in the fast exchange and the amide bond in the fast rotation on the NMR time scale. Meanwhile, there are no significant upfield chemical shifts caused by elevated temperatures. In a high concentration (15 mM), both NH and amide protons appears as broad peaks below −30°C (243 K). With increasing temperatures, the NH proton peak disappears while the two amide proton peaks become much sharper and shift systematically upfield (Figure 5b). The VT NMR studies reveal a dynamic slowdown of the tautomerization and rotation with the increased concentration.
Figure 4.
Molecular orbital energies (eV), dipole moments (μ, D), and torsion angles (deg) for the two tautomers of 5c in chloroform from the DFT calculation. The two tautomers of 5c are designated according to nomenclature of amino acids and peptides recommended by IUPAC and IUBMB (http://www.chem.qmul.ac.uk/iupac/index.html) for histidine.
Scheme 3.
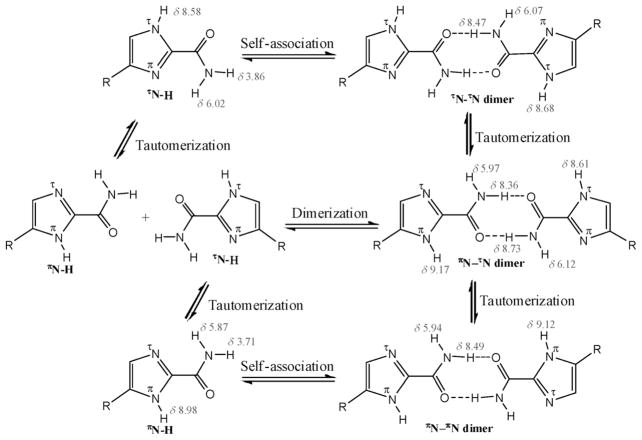
Chemical equilibria of 4(5)-SiOM-1-H-imidazole-2-carboxamide in chloroform (numbers in red: calculated chemical shifts (ppm) of protons in 5c dimers).
Figure 5.
VT NMR spectra of 5c in CDCl3 at a concentration of a) 2 mM and b) 15 mM.
To understand these NMR data, we propose a scheme to depict the chemical equilibriums of 1-H-imidazole-2-carboxamide taking place in chloroform (Scheme 3). The 1-H-imidazole-2-carboxamide molecule can spontaneously undergo both tautomerization within the imidazole ring and dimerizations (or self associations) between the amides through hydrogen bonding interactions. It has been reported that the tautomerization rate decreased with an increase in the size of an N-substituent on carboxamide at the 2 position of imidazole, presuming that the formation of tautomers was through an intermolecular proton exchange mechanism.[24] Although the intermolecular proton transfer in nonprotic solvents is an energetically favorable process in the prototropic tautomerization of imidazole, an intramolecular proton transfer is also feasible in imidazole,[25] supported by low-temperature IR experiments in inert gas matrices that indicate the existence of two imidazole tautomers in isolated adenine and guanine.[26] Since our NMR data indicate that the NH proton was not involved in hydrogen bonding, we believe that an intramolecular mechanism may play a major role in tautomerization of 1-H-imidazole-2-carboxamide. The amide group functions as a carrier for transportation of the proton from one nitrogen atom to another by rotating around the imidazole plane. To do so, the NH proton first migrates to the nearby oxygen of amide, and then the protonated amide turns 180° to other side of the imidazole ring and releases the proton to the nearby nitrogen. Thus, formation of a hydrogen bonded dimer slows down the rotation of the amide and reduces the exchange rate of the proton, resulting in the broadening (or disappearing into the baseline) of the NH proton signal in NMR spectra. The signal reappears when the temperature continues lowering to further slow down the proton exchange. That is a phenomenon commonly observed in dynamic NMR. As shown in Scheme 3, there are three different dimers that can be formed from two tautomers. Our DFT calculation shows that there are no significant differences in total energy and dimerization energy among these dimers (Table S1 in the Supporting Information), which implies that there is no preference to form these dimers. The calculated chemical shifts of the hydrogen bonded amide proton and the NH proton are close to each other (see Scheme 3) so that these dimers may not readily be distinguished by routine NMR experiments and the NH proton may coalesce with the hydrogen bonding amide proton. That requires more sophisticated NMR techniques, such as dynamic NMR at very low temperatures, to identify these individual dimers.[27] Through NMR dilution experiments, the self association constant of 5c was determined (Figure S4 in the Supporting Information). The curve fitting was carried out in the computer program HypNMR 2008. The first attempt to fit data from the hydrogen bonded amide proton into a dimerization (R2) model was failed. However, the data from the non hydrogen bonded amide proton fit into the R2 model convergently, giving the constant 2.0 (1) (in log β). To explain the non-convergence of the hydrogen bonded amide proton, we deduce that its chemical shifts in the lower concentrations were coalesced with those of the NH proton, based on observation that the amide peak were broadened out and shifted downfield with decreasing concentrations (Figure S4 in the Supporting Information).
As far as physical properties are concerned, two tautomers of 5c have relatively small dipole moments (Figure 4), compared with DNA bases (G: 6.55 D> C: 6.39 D> T: 4.31 D> A: 2.56 D).[28] This may make it less effective in the base stacking. The HOMO energy of the 1-H-imidazole-2-carboxamide molecule (6.47 eV for τN(H) tautomer and −6.11 eV for πN(H) tautomer) is higher than those of DNA bases (G: −7.75 eV; A: −8.24 eV; C: −8.87 eV; T: −9.14 eV), closer to the Fermi level of gold (−5 to −5.31 eV);[29] however, its HOMO–LUMO gap (Figure 4) is much larger than those of DNA bases (3–4 eV ). Thus, 1-H-imidazole-2-carboxamide may be more feasible for charge transfer than the DNA bases, while less effective for electron transfer due to its higher LUMO energy (−0.77 for τN(H) tautomer and −0.50 eV for πN(H) tautomer).
Interactions of 1-H-imidazole-2-carboxamide with DNA bases
We have studied the hydrogen bonding interactions of 5c with DNA bases by NMR titration in chloroform. Four naturally occurring nucleosides (referred as to dA, dC, dG, and dT) were used as the model compounds of the bases imbedded in a single stranded DNA, which were made soluble in chloroform by silylating their hydroxyl groups with tert-butyldimethylsilyl chloride (TBDMSCl),[30] and 5c was only used as a substrate of the titration due to its limited solubility in chloroform, the concentration of which was held constant at 5 mM so that it existed mainly in a monomer form. It was found that the chemical shift of the NH proton changed irregularly with concentrations of the titrants and the analyzable data were obtained from monitoring changes in chemical shifts of the amide protons. The control experiments were titrating dT with dA and dG. The program HypNMR 2008 was employed to determine equilibrium constants from the titration data. First, stoichiometries of the interactions between 5c and nucleosides were estimated by plotting the NMR titrating data as nucleoside to 5c mole ratios versus chemical shifts of the amide proton (Figure S5 in the Supporting Information). From the mole ratio plots, we deduce that 5c may form 1:1 complexes with the nucleoside dA and dT, and 1:2 complexes with nucleoside dC and dG. Thus, the associations between 5c (S) and the nucleosides (N) can be expressed in multistep equilibriums as shown below:
Based on this thermodynamic process, the titration data were then fit into the 1:1, 1:2, or 1:1/1:2 binding isotherm by nonlinear regression in the HypNMR program. It was found that the data from titrating with dA and dT were fit into the 1:1 model and those from titrating with dC and dG were fit into the 1:1/1:2 model convergently (Figure S6 in the Supporting Information). The association constants derived from the curving fitting are listed in Table 1. Following the same procedure just mentioned above, we determined the association constant of the Watson–Crick base pair dA–dT, which is close to the value reported in literature,[31] and the association constant of a mismatched base pair dG–dT, which is consistent with one derived from curving fitting in the program Origin 8 (Log β11 = 2.52). These indicate that our data analysis process is reasonably reliable. From the association constants, we see that the nucleosides form the 1:1 complexes with 5c in a tendency of dG> dC≫dT> dA in chloroform. Furthermore, dG and dC can also form 2:1 complexes with 5c, which are more stable than the 1:1 complexes. The data reported here demonstrate that 5c can interact with the nucleosides through their nucleobases in the organic phase although the interactions do not, in themselves, represent strong recognition. We predict that the 1-H-imidazole-2-carboxamide molecules tethered to two electrodes should form a more stable 2:1 complex with each of the DNA bases in a nanogap than its 1:1 counterpart in the solution. In our previous study, we found that bond lifetimes with DNA bases were greatly enhanced when the benzamide molecules were tethered in a nanogap for the recognition.[12] We recently observed the same phenomenon in a simpler case of a nucleobase recognizing its complement in a nanogap.[32] Since 1-H-imidazole-2-carboxamide interacts with each of DNA bases differently, the DNA bases should be electronically discriminable in the nanogap functionalized with the reading molecules.
Table 1.
Association constants of 5c with nucleosides determined from curve fitting of NMR titration data.
| Log β11 | Log β12 | |
|---|---|---|
| 5c-dA | 1.08(9) | – |
| 5c-dC | 3.1(1) | 5.04(9) |
| 5c-dG | 3.31(2) | 5.49(1) |
| 5c-dT | 1.46(5) | – |
| dA-dT | 1.62(1) | – |
| dG-dT | 2.56(6) | – |
Conclusion
To complement our recognition tunneling technique, we have designed 1-H-imidazole-2-carboxamide as a potential universal reader to read all of the possible individual DNA bases in a single stranded DNA. A synthetic route has been developed for introducing a linkage to the 4(5)-position of 1-H-imidazole-2-carboxamide. Our data show that 1-H-imidazole-2-carboxamide is suitable to work under the physiological conditions. In the present study, we demonstrate by NMR titration that the 1-H-imidazole-2-carboxamide molecule can form hydrogen bonded complexes with all four naturally occurring DAN bases in the dielectric and aprotic environments. The recognition of DNA bases by the imidazole molecules immobilized in nanogaps is the subject of a manuscript in preparation.
Experimental Section
General information
All reagents were obtained from commercial suppliers unless otherwise stated. Where necessary, organic solvents were routinely dried and/or distilled prior to use and stored over molecular sieves under nitrogen. All reactions requiring anhydrous conditions were performed under a nitrogen atmosphere. Reactions were monitored by TLC on silica gel plates. Flash chromatography was performed in an automated flash chromatography system (CombiFlash Rf, Teledyne Isco, Inc.) with silica gel columns (60–120 mesh). Proton NMR (1H) spectra were recorded on a Varian 400 MHz spectrometer or a Varian 500 MHz spectrometer. 1H chemical shifts in chloroform were referenced relative to deuterated solvents (CDCl3: δH = 7.26 ppm, CD3OD: δH = 3.31 ppm, and DMSO: δH = 2.50 ppm). 2D NOE were recorded at 400 MHz on a Bruker 400 MHz spectrometer. In VT NMR, temperature was calibrated with a standard solution of 100% CH3OH and regulated to an accuracy of −0.1°C by a Eurotherm Variable Temperature Unit on the Bruker NMR or a Highland Technologies Temperature unit on the Varian NMR System. Below zero Celsius temperatures were achieved with a Liquid Nitrogen Heat Exchanger on the Bruker and FTS Cooling System (Stone Ridge, New York) on the Varian. HRMS spectra were recorded using the atmospheric pressure chemical ionization (APCI) technique. Mettler Toledo Portable Meter was used for pH determination. UV spectra were recorded on a Varian Cary 300 UV spectrophotometer. Mathematical analysis of NMR data and graphic presentation of results were performed using the program HypNMR2008, purchased from Protonic Software, an upgraded release of HYPNMR handling general host-guest association equilibria.[33]
4(5)-(2-(Benzylthio)ethyl)-1-H-imidazole (1a)
Sodium hydride (60% in mineral oil, 810 mg, 20.2 mmol) was added to a solution of 4(5)-(2-bromoethyl)-1-H-imidazole hydrochloride and 4(5)-(2-chloroethyl)-1-H-imidazole hydrochloride[34] (3.32 g) in DMF (30 mL) at 0°C under nitrogen. The resulting mixture was stirred for 30 min, followed by adding a solution of benzyl mercaptan (5.05 g, 40.6 mmol) in DMF (30 mL) dropwise. The mixture was stirred at 0°C for 2 h, and then allowed to warm to room temperature and stirred overnight. The solvent was removed by rotary evaporation under reduced pressure, and then the residue was suspended in water (100 mL), extracted with chloroform (4050 mL). The combined organic layer was washed with water (3050 mL), brine (100 mL), and then concentrated. The crude product was purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:10 to 10:100. Compound 1a was obtained as a white powder (605 mg). 1H NMR (400 MHz, CDCl3): δ = 2.73 (t, 2H, 3J (H,H)=7.2 Hz), 2.86 (t, 2H, 3J (H,H)=7.2 Hz), 3.72 (s, 2H), 6.79 (s, 1H), 7.23–7.31 (m, 5H), 7.54 ppm (s, 1H); HRMS (APCI+): m/z calculated for C12H14N2S+H: 219.0956; found: 219.0962.
1-Benzyl-4(5)-(2-(benzylthio)ethyl)-1H-imidazole (2a)
Sodium hydride (60% in mineral oil, 20 mg, 0.5 mmol) was added to a solution of 4(5)-(2-(benzylthio)ethyl)-1-H-imidazole (1a) (109 mg, 0.5 mmol) in dioxane (2 mL) at 0°C under nitrogen. The resulting mixture was stirred for 30 min, and then the benzyl bromide (77 mg, 0.45 mmol) was added dropwise. The mixture was heated to 90°C and stirred overnight. After cooling to room temperature, the reaction was quenched with water (5 mL) and extracted with chloroform (505 mL). The combined organic layer was washed with brine (10 mL) and concentrated. The crude product was purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 8:100. Compound 2a was obtained as clear oil (129 mg, 84%). 1H NMR (400 MHz, CDCl3): δ = 2.72 (m, 2H), 2.80 (m, 2H), 3.61 and 3.69 (s, s, 2H), 4.98 and 5.02 (s, s, 2H), 6.61 and 6.82 (s, s, 1H), 7.43 and 7.44 (s, s, 2H), 7.12–7.36 ppm (m, 10H);HRMS (APCI+): m/z calculated for C19H20N2S+H: 309.1425.1; found:309.1420.
1-Benzyl-4(5)-(2-(benzylthio)ethyl)-1H-imidazole-2-carbonitrile (3a)
Cyanogen bromide (111 mg, 1.05 mmol) was added to a solution of 4-dimethylaminopyridine (128 mg, 1.05 mmol) in DMF (1.5 mL) at 0°C under nitrogen. After 30 min, a solution of 2a (129 mg, 0.42 mmol) in DMF (0.6 mL) was added dropwise to the CAP solution at 0°C. The resulting mixture was stirred at 40°C for 16 h and then quenched with sat. sodium bicarbonate solution (10 mL), and extracted with ethyl acetate (405 mL). The combined organic layer was concentrated, and the residue was purified by flash chromatography in a silica gel column with a gradient of ethyl acetate in hexanes from 0:100 to 20:100. Compound 3a was obtained as yellowish oil (117 mg, 84%). 1H NMR (400 MHz, CDCl3): δ=2.49–2.78 (m, 4H), 3.61 and 3.67 (s, s, 2H), 5.16 and 5.18 (s, s, 2H), 6.77 and 6.96 (s, s, 1H), 6.99–7.38 ppm (m, 10H); IR (KBr): ṽ =2201 cm−1 (CN stretching); HRMS (APCI+): m/z calculated for C20H19N3S+H: 334.1378; found: 334.1366.
1-Benzyl-4(5)-(2-(benzylthio)ethyl)-1-H-imidazole-2-carboxamide (4a)
A mixture of 3a (48 mg, 0.14 mmol) in sulfuric acid (20% by volume, 0.3 mL) and TFA (167 uL) was stirred overnight at 75°C. After cooling, the solvent was removed by rotary-evaporation, and the residue was purified by flash chromatography in a silica gel column with a gradient of ethyl acetate in hexanes from 0:100 to 20:100. Compound 4a was obtained as a white powder (23 mg, 46%). 1H NMR (400 MHz, CDCl3): δ=2.44 (t, 2H, 3J (H,H)=7.6 Hz), 2.59 (t, 2H, 3J (H,H)=7.6 Hz), 3.55 (s, 2H), 5.63 (s, 2H), 6.78 (s, 1H), 6.92–7.24 ppm (m, 12H); HRMS (APCI+): m/z calculated for C20H21N3OS+H: 352.1484; found: 352.1493.
4(5)-(2-Mercaptoethyl)-1-H-imidazole-2-carboxamide (5a)
A solution of 4a (34 mg, 0.097 mmol) in diethyl ether (0.2 mL) was added to a solution of sodium (40 mg) in ammonia (anhydrous) at − 76°C under nitrogen. During the reaction time, more sodium was added to keep the solution blue. After 5 h, the reaction was quenched by addition of ammonium chloride (40 mg), allowing ammonia to evaporate at room temperature. The residue was washed with hexanes and diethyl ether, and separated by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 50:50. Compound 5a was obtained as a white powder (15 mg, 77%): 1H NMR (400 MHz, CD3OD): δ = 3.04–3.15 (m, 4H), 4.14 (s, 1H), 7.32 ppm (s, 1H); HRMS (APCI+): m/z calculated for C6H9N3OS+H: 172.0545; found: 172.0522.
N-Phthaloyl histamine (1b)
The compound was synthesized following a procedure reported in literature.[35] Finely powdered N-ethoxycarbonylphthalimide (12.5 g, 55 mmol) was added to a vigorously stirred solution of histamine dihydrochloride (9.2 g, 50 mmol) and sodium carbonate (10.6 g, 100 mmol) in water (250 mL) at room temperature in a period of 30 min. After the addition, the mixture was stirred for an additional hour, filtered, and the product recrystallized from aqueous ethanol to give compound 1b as a white power (9.2 g, 76%). 1H NMR (400 MHz, DMSO): δ=2.93 (t, 2H, 3J (H,H)=6.8 Hz), 3.90 (t, 2H, 3J (H,H)= 6.8 Hz), 6.76 (s, 1H), 7.46 (s, 1H), 7.74–7.82 ppm (m, 4H).
1-Benzyl-4(5)-(2-(phthaloyl)ethyl)-1-H-imidazole (2b)
Sodium hydride (60% in mineral oil, 200 mg, 5 mmol) was added to a solution of N-Phthaloylhistamine 1b (1.21 g, 5 mmol) in dioxane (10 mL) at 0°C under nitrogen. The resulting mixture was stirred for 30 min, followed by adding a solution of benzylbromide (0.81 g, 4.75 mmol) in dioxane (5 mL) dropwise. The mixture was stirred overnight at room temperature, quenched with water (20 mL), and extracted with chloroform (30 20 mL). The combined organic layer was washed with water (3050 mL), brine (50 mL), and concentrated. The crude product was purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 10:100. The compound 2b was obtained as yellowish oil (1.56 g, 94%). 1H NMR (400 MHz, CDCl3): δ = 2.96 (t, 2H, 3J (H,H)=7.4 Hz), 3.98 (t, 2H, 3 J (H,H)=7.4 Hz), 5.02 (s, 1H), 6.68 (s, 1H), 7.10–7.30 (m, 5H), 7.44 (s, 1H), 7.68–7.81 ppm (m, 4H)
1-Benzyl-4(5)-(2-(phthaloyl)ethyl)-1-H-imidazole-2-carbonitrile (3b)
Cyanogen bromide (2.48 g, 23.5 mmol) was added to a solution of 4-di-methylaminopyridine (2.86 g, 23.5 mmol) in DMF (23 mL) at 0°C under nitrogen. After 30 min, a solution of 2b (1.56 g, 4.7 mmol) in DMF (10 mL) was added dropwise to the CAP solution. The resulting mixture was stirred at 40°C for 16 h, quenched with sat. sodium bicarbonate solution (50 mL), and then extracted with ethyl acetate (4050 mL). The combined organic layer was concentrated, and purified by flash chromatography in a silica gel column with a gradient of ethyl acetate in hexanes from 0:100 to 50:50. Compound 3b was obtained as yellowish oil (1.17 g, 70%). 1H NMR (400 MHz, CDCl3): δ = 2.91 (t, 2H, 3J (H,H)=7.0 Hz), 3.95 (t, 2H, 3J (H,H)=7.0 Hz), 5.20 (s, 2H), 6.89 (s, 1H), 7.09–7.36 (m, 5H), 7.69–7.82 ppm (m, 4H); IR (KBr): ṽ =2230 cm−1 (CN stretching); HRMS (APCI+): m/z calculated for C21H18N4O3+H: 357.1351; found: 357.1359.
1-Benzyl-4(5)-(2-(amine)ethyl)-1-H-imidazole-2-carboxamide (4b)
A solution of 3b (166 mg, 0.47 mmol) in methanol (13 mL) was basified to pH ~9 by addition of NH3·H2O (28–30%), to which H2O2 (30%, 1.25 mL) was added dropwise. The resulting mixture was stirred overnight at room temperature, and then concentrated. The residue was dissolved in hydrazine (1.0M in THF, 2.2 mL) and refluxed overnight. After removing the solvent, the residue was purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 50:100. Compound 4b was obtained as a white powder (77 mg, 32%). 1H NMR (400 MHz, CD3OD): δ = 2.89 (t, 2H, 3J (H,H)= 6.6 Hz), 3.24 (t, 2H, 3J (H,H)=6.6 Hz), 5.69 (s, 2H), 7.18 (s, 1H), 7.26–7.46 ppm (m, 5H); HRMS (APCI+): m/z calculated for C13H16N4O+H: 245.1402; found: 245.1402.
4(5)-(2-(Amine)ethyl)-1-H-imidazole-2-carboxamide (5b)
A solution of 4b (40 mg, 0.16 mmol) in diethyl ether (0.2 mL) was added to a solution of sodium (40 mg) in ammonia (anhydrous) at − 76°C under nitrogen. During the reaction time, more sodium was added to keep the solution blue. After 5 h, the reaction was quenched with ammonium chloride (40 mg), and then the ammonia was allowed to evaporate at room temperature. The residue was wished with hexanes and diethyl ether, and purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 50:50. Compound 5b was obtained as a white powder (21 mg, 85%). 1H NMR (400 MHz, CD3OD): δ=2.98 (t, 2H, 3J (H,H)=6.6 Hz), 3.26 (t, 2H, 3J (H,H)=6.6 Hz), 7.13 (s, 1H), 7.37 ppm (brs, 2H); HRMS (APCI+): m/z calculated for C6H10N4O+H: 155.0933; found: 155.0929.
4(5)-((tert-butyldimethylsilyloxy)methyl)-1-H-imidazole (1c)
The compound was synthesized following a procedure reported in literature.[36] To a solution of 4(5)-hydroxymethyl-1-H-imidazole hydrochloride (200 mg, 1.48 mmol) in DMF (3 mL) were successively added imidazole (334 mg, 4.90 mmol), TBDMSCl (246 mg, 1.64 mmol) and 4-dimethylaminopyridine (36.4 mg, 0.30 mmol). The resulting mixture was stirred at room temperature overnight and then at 60°C for 3 h, diluted with water (2 mL), and then extracted with a co-solvent of hexanes and ether (v/v= 1:1, 6 mL). The organic layer was washed with water, brine, and then concentrated. Compound 1c was obtained as a colorless solid (250 mg, 79%). 1H NMR (400 MHz, CDCl3): δ = 0.09 (s, 6H), 0.92 (s, 9H), 4.74 (s, 2H), 6.92 (s, 1H), 7.60 ppm (s, 1H).
1-Benzyl-4(5)-((tert-butyldimethylsilyloxy)methyl)-1-H-imidazole (2c)
Sodium hydride (60% in mineral oil, 88 mg, 2.2 mol) was added to a solution of 1c (0.47 g, 2.2 mmol) in dioxane (4.2 mL) at room temperature. After 30 min, benzyl bromide (248 μL, 2.09 mmol) was added into the mixture, which was stirred at 90°C for 18 h and then the solvent was removed by rotary evaporation under reduced pressure. The residue was diluted with water (5 mL) and extracted with chloroform (305 mL). The combined organic layer was washed with water (305 mL), brine (10 mL), and then concentrated. The crude product was purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 10:100. Compound 2c was obtained as yellowish oil (0.44 g, 66%). 1H NMR (400 MHz, CDCl3): δ = 0.09 (s, 6H), 0.86 and 0.91 (s, s, 9H), 4.52 and 4.69 (s, s, 2H), 5.06 and 5.20 (s, s, 2H), 6.82–7.46 ppm (m, 6H).
1-Benzyl-4(5)-((tert-butyldimethylsilyloxy)methyl)-1-H-imidazole-2-carbonitrile (3c)
Cyanogen bromide (132 mg, 1.25 mmol) was added to a solution of 4-dimethylaminopyridine (152 mg, 1.25 mmol) in DMF (2.5 mL) at 0°C under nitrogen. After 30 min, a solution of 2c (151 mg, 0.5 mmol) in DMF (0.6 mL) was added dropwise to the CAP solution. The resulting mixture was stirred at 40°C for 16 h, quenched with sat. sodium bicarbonate solution (14 mL), and extracted with ethyl acetate (3010 mL). The combined organic layer was concentrated, and the residue was purified by flash chromatography on a silica gel column with a gradient of ethyl acetate in hexanes from 0:100 to 20:100. Compound 3c was obtained as yellowish oil (133 mg, 85%). IR: v=2240 cm−1 (CN stretching); 1H NMR (400 MHz, CDCl3): δ = 0.08 (s, 6H), 0.88 (s, 9H), 4.66 (s, 2H), 5.25 (s, 2H), 6.99 (s, 1H), 7.24–7.40 ppm (m, 5H); HRMS (APCI+): m/z calculated for C18H25N3OSi+H: 328.1845; found: 328.1847.
1-benzyl-4(5)-((tert-butyldimethylsilyloxy)methyl)-1-H-imidazole-2-carboxamide (4c)
A solution of 3c (133 mg, 0.41 mmol) in methanol (10 mL) was basified to pH 9–10 by addition of ammonium hydroxide. To this solution, H2O2 (30%, 1.25 mL) was added dropwise at 0 °C. The resulting mixture was stirred for 16 h at room temperature. After the solvent was removed, the residue was purified by flash chromatography in a silica gel column with a gradient of ethyl acetate in hexanes from 0:100 to 50:100. Compound 4c was obtained as a white powder (90 mg, 64%). 1H NMR (400 MHz, CDCl3): δ = 0.08 (s, 6H), 0.90 (s, 9H), 4.53 and 4.64 (s, s, 2H), 5.26 (brs, 1H), 5.68 and 5.87 (s, s, 2H), 6.92 and 6.99 (s, s, 1H), 7.08–7.33 ppm (m, 6H); HRMS (APCI+): m/z calculated for C18H27N3O2Si+H: 346.1951; found: 346.1949.
4(5)-((tert-Butyldimethylsilyloxy)methyl)-1-H-imidazole-2-carboxamide (5c)
A solution of 4c (78 mg, 0.23 mmol) in diethyl ether (12 mL) was added to a solution of sodium (80 mg) in ammonia at −76°C under nitrogen. During the reaction time, more sodium was added to keep the solution blue. After 2 h, the reaction was quenched with ammonium chloride (80 mg), and the ammonia allowed evaporating at room temperature. The residue was washed with hexanes and diethyl ether, and purified by flash chromatography in a silica gel column with a gradient of methanol in dichloromethane from 0:100 to 50:100. Compound 5c was obtained as a white powder (18 mg, 35%). 1H NMR (400 MHz, CDCl3): δ = 0.10 (s, 6H), 0.91 (s, 9H), 4.73 (s, 2H), 5.93 (brs, 1H), 7.08 (s, 1H), 7.64 pm (brs, 1H); HRMS (APCI+): m/z calculated for C11H21N3O2Si+H: 256.1481; found: 256.1488.
UV determination of acid dissociation constants (pKa)
for the pKa determination, a set of UV spectra were first collected at different pHs, from which data was then extracted and analyzed according to the method reported in literature.[37] The pKa values of 1-H-imidazole-2-carboxamide were determined from Method a, and those of 5a from both Method a and b.
Method a
14 buffer solutions with different pH values were prepared using KCl/HCl (pH 1), C6H8O7/HCl/NaCl (pH 2), C6H8O7/NaOH/NaCl (pH 3), C6H8O7/NaOH/NaCl (pH 4), C6H8O7/NaOH (pH 5, 6), KH2PO4/ Na2HPO4 (pH 7), Na2B4O7/HCl (pH 8, 9), Na2B4O7/NaOH (pH 10), H3BO3/NaOH/KCl (pH 11), Na2HPO4/NaOH (pH 12), NH2CH2COOH/NaCl/NaOH (pH 13), NaOH (pH 14). Stock solutions of 1-H-imidazole-2-carboxamide and 4(5)-(2-mercaptoethyl)-1-H-imidazole-2-carboxamide (5a) were prepared in DI water with concentration of 1.65010−3M and 7.53010−4M, respectively. Working solutions were prepared by 20-fold dilution of the stock solutions with buffers. Measurements were carried out in the same day as fresh working solutions were prepared.
Method b
An aliquot (0.6 mL) of a 5a stock solution (2010−3M) in aq. HCl (0.1M) was added to a solution of 5a (2 X 10−3M) in aq. NaOH (0.1M), followed by recording the UV spectrum and measuring the pH value of the resultant solution. The same procedure was repeated in the same solution until a set of UV spectra were collected with the pH ranging from1.0 to 13.0. The temperature was controlled between 25 to 27°C.
Data analysis
The pKas were determined based on the Henderson-Hasselbalch equation, which can be expressed as below:
where β is the degree of dissociation.
Given that UV absorption is additive, the total absorbance of a weak acid solution can be expressed as the sum of those of the acid and its conjugate base. The UV absorbances of the imidazole-2-carboxamide at the wavelength of 258 nm (Aλ1), 268 nm (Aλ2), and 281 nm (Aλ3) are presented in following equations. CI, CII, CIII are concentrations of the form I, II, III, respectively, and b is the light path length. Accordingly, the extinction coefficients of I, II, III at 258 nm are designated as ε1–I, ε1–II, and ε1–III for example.
Assuming that the imidazole exists in a positively charged form (I) at pH 1.7, neutral form (II) at pH 8.3, and negatively charged form (III) at pH 12.2, the extinction coefficient of each form at its absorption maximum was estimated from the UV absorbance (Table 2). Using these equations, the degrees of dissociation (β) were calculated from the total absorbances at different pHs with the total concentration of the species constant. The β values were plotted against corresponding pHs, which was fit with a sigmoidal function (Boltzmann fitting) in the Origin software. The pKa value was directly read out from pH at β = 0.5 on the fitting curve. To simplify the calculation, CIII was set to zero at pH lower than 7.0 and CI was set to zero at pH higher than 7.0. The distribution of three forms of 5a at different pHs was calculated in the same manner, shown in Figure 2c.
Table 2.
Extinction coefficients (ε, mM−1cm−1) of three forms of 5a at their maximum wavelengths.
| ε258 [nm] | ε268 [nm] | ε281 [nm] | |
|---|---|---|---|
| I | 6.761 | 5.481 | 1.816 |
| II | 6.039 | 7.231 | 4.908 |
| III | 3.796 | 5.697 | 7.158 |
Computational methods
All the DFT calculations were performed in the Spartan′10 for Windows, Wavefunction Inc. Individual 2D chemical structures were drawn in ChemBioDraw Ultra 11 and exported to Spartan′10 to generate corresponding 3D structures from which hydrogen bonding dimers were generated as well. The geometries of these structures were subjected to energy minimization using the built-in MMFF molecular mechanics prior to the DFT calculations. All the structures were first calculated using the B3LYP with the 6–311+ +G** basis set in vacuum, followed by B3LYP/6–31G* in chloroform, from which NMR chemical shifts were simultaneously calculated. The DFT calculations for hydrogen bonding interactions of the imidazole-2-carboxamide with DNA bases were performed using B3LYP/6–31G* basis set in vacuum.
1H NMR titrations
1H NMR titration experiments were carried out at 24±1°C using either a Varian 500 spectrometer operating at 500 MHz or a Varian 400 spectrometer operating at 400 MHz. All chemical shifts are reference to CDCl3 and reported in ppm. The 5c solution with a known concentration, typically 5010−3M, was made up in an NMR tube using deuterated chloroform (0.5 mL). The titrating solutions of TBDMS silylated nucleosides were made up in volumetric flasks (2 mL) with a concentration much greater than that of 5c. In a typical titration, one of the titrating solutions was added to the 5c solution in 7–14 separate portions. Following each of additions, NMR spectrum was recorded. For example, the dT nucleoside solution with the concentration of 800010−3M was added incrementally in the order of 1.25, 1.25, 1.25, 1.25, 2.5, 2.5, 2.5, 2.5, 5, 5, and 5 μL aliquots, up to a total volume of 30 μL. The titration data were analyzed by nonlinear regression analysis using the HypNMR2008 program. The stoichiometry of 5c-nucleoside complexes was established by mole ratio plots.[38]
Acknowledgments
This project is funded by the DNA sequencing technology program of the National Institute of Human Genome Research (HG004378). We thank Dr. Brian Cherry of the magnetic resonance research center at ASU for his technical support and informative discussion during the NMR experiments.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/chem.201103306.
Contributor Information
Prof. Stuart Lindsay, Email: Stuart.Lindsay@asu.edu.
Prof. Peiming Zhang, Email: peiming.zhang@asu.edu.
References
- 1.Chang S, He J, Zhang P, Gyarfas B, Lindsay S. J Am Chem Soc. 2011;133:14267–14269. doi: 10.1021/ja2067737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yngvadottir B, MacArthur DG, Jin H, TylerSmith C. GenomeBiology. 2009;10:237231–237234. [Google Scholar]
- 3.Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukhopadhyay R. Anal Chem. 2009;81:1736–1740. doi: 10.1021/ac802712u. [DOI] [PubMed] [Google Scholar]
- 5.Alkan C, Sajjadian S, Eichler EE. Nat Methods. 2011;8:61–65. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Snyder M, Du J, Gerstein M. Genes Dev. 2010;24:423–431. doi: 10.1101/gad.1864110. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Venter JC. Nature. 2010;464:676–677. doi: 10.1038/464676a. [DOI] [PubMed] [Google Scholar]
- 7.a) Zwolak M, Ventra MD. Rev Mod Phys. 2008;80:141–165. [Google Scholar]; b) Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Ventra MD, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Nat Nanotechnol. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 9.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Proc Natl Acad Sci USA. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasianowicz JJ, ERIC EB, Branton D, Deamer DW. Proc Natl Acad Sci USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Zwolak M, Ventra MD. Nano lett. 2005;5:421–424. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]; b) Ivanov AP, Instuli E, McGilvery C, Baldwin G, McComb DW, Albrecht T, Edel JB. Nano lett. 2011;11:279–285. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang S, He J, Chang S, Zhang P, Liang F, Li S, Tuchband M, Fuhrman A, Ros R, Lindsay S. Nat Nanotechnol. 2010;5:868–873. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam SW, Rooks MJ, Kim KB, Rossnagel SM. Nano Lett. 2009;9:2044–2048. doi: 10.1021/nl900309s. [DOI] [PubMed] [Google Scholar]
- 14.Richaud A, Barba-Behrens N, Mendez F. Org Lett. 2011;13:972–975. doi: 10.1021/ol103011h. [DOI] [PubMed] [Google Scholar]
- 15.Bergstrom DE, Zhang P, Zhou J. J Chem Soc Perkin Trans. 1994;1:3029–3034. [Google Scholar]
- 16.Kirk KL. J Org Chem. 1978;43:4381–4383. [Google Scholar]
- 17.McCarthy JR, Matthews DP, Whitten JP. Tetrahedron Lett. 1985;26:6273–6276. [Google Scholar]
- 18.Moorthy JN, Singhal N. J Org Chem. 2005;70:1926–1929. doi: 10.1021/jo048240a. [DOI] [PubMed] [Google Scholar]
- 19.Bergstrom DE, Zhang P. Tetrahedron Lett. 1991;32:6485–6488. [Google Scholar]
- 20.Gilchrist TL. Heterocyclic Chemistry. 3. Longman; 1997. [Google Scholar]
- 21.Homer RB, Johnson CD. In: The Chemistry of amides. Zabicky J, editor. Wiley; 1970. pp. 187–243. [Google Scholar]
- 22.Roig B, Chalmin E, Touraud E, Thomas O. Talanta. 2002;56:585–590. doi: 10.1016/s0039-9140(01)00580-x. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn B, Mohr P, Stahl M. J Med Chem. 2010;53:2601–2611. doi: 10.1021/jm100087s. [DOI] [PubMed] [Google Scholar]
- 24.Papadopoulos EP, Hollstein U. Organic Magnetic Resonance. 1982;19:188–191. [Google Scholar]
- 25.a) Ten GN, Baranov VI. Journal of Applied Spectroscopy. 2008;75:168–173. [Google Scholar]; b) Ten GN, Burova TG, Baranov VI. Journal of Structural Chemistry. 2007;48:623–633. [Google Scholar]
- 26.a) Stepanian SG, Sheina GG, Radchenko ED, Blagoi YP. Journal of Molecular Structure. 1985;131:333–346. [Google Scholar]; b) Sheina GG, Stepanian SG, Radchenko ED, Blagoi YP. Journal of Molecular Structure. 1987;158:275–292. [Google Scholar]
- 27.Weisz K, J3hnchen J, Limbach H-H. J Am Chem Soc. 1997;119:6436–6437. [Google Scholar]
- 28.Sponer J, Leszczynski J, Hobza P. Biopolymers. 2002;63:3–31. doi: 10.1002/1097-0282(2001)61:1<3::AID-BIP10048>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Apalkov V, Wang X-F, Chakraborty T. In: Charge Migration in DNA. Chakraborty T, editor. Springer; 2007. pp. 77–119. [Google Scholar]
- 30.Chang S, Huang S, He J, Liang F, Zhang P, Li S, Chen X, Sankey O, Lindsay S. Nano Lett. 2010;10:1070–1075. doi: 10.1021/nl1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sartorius J, Schneider HJ. Chem Eur J. 1996;2:1446–1452. [Google Scholar]
- 32.Fuhrmann A, Getfert S, Fu Q, Reimann P, Lindsay S, Ros R. Biophysical Journal. 2011 doi: 10.1016/j.bpj.2012.04.006. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.a) Frassineti C, Alderighi L, Gans P, Sabatini A, Vacca A, Ghelli S. Anal Bioanal Chem. 2003;376:1041–1052. doi: 10.1007/s00216-003-2020-0. [DOI] [PubMed] [Google Scholar]; b) Frassineti C, Ghelli S, Gans P, Sabatini A, Moruzzi MS, Vacca A. Anal Biochem. 1995;231:374–382. doi: 10.1006/abio.1995.9984. [DOI] [PubMed] [Google Scholar]
- 34.Ting R, Lermer L, Perrin DM. J Am Chem Soc. 2004;126:12720–12721. doi: 10.1021/ja046964y. [DOI] [PubMed] [Google Scholar]
- 35.Emmett JC, Holloway FH, Turner JL. J Chem Soc, Perkin Trans. 1979;1:1341–1344. [Google Scholar]
- 36.Amino Y, Eto H, Eguchi C. Chemical & Pharmaceutical Bulletin. 1989;37:1481–1487. doi: 10.1248/cpb.37.506. [DOI] [PubMed] [Google Scholar]; Pharmaceutical Bulletin. 1989;37:1481–1487. [Google Scholar]
- 37.Rether C, Schmuck C. Eur J Org Chem. 2011:1459–1466. [Google Scholar]
- 38.Cheng PN, Lin CF, Liu YH, Lai CC, Peng SM, Chiu SH. Org Lett. 2006;8:435–438. doi: 10.1021/ol052679n. [DOI] [PubMed] [Google Scholar]